Upstream CtrA-binding sites both induce and repress pilin gene expression in Caulobacter crescentus
- PMID: 39030481
- PMCID: PMC11264516
- DOI: 10.1186/s12864-024-10533-6
Upstream CtrA-binding sites both induce and repress pilin gene expression in Caulobacter crescentus
Abstract
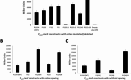
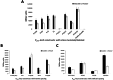
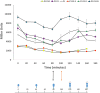
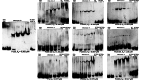
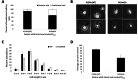
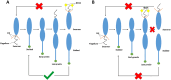
Pili are bacterial surface structures important for surface adhesion. In the alphaproteobacterium Caulobacter crescentus, the global regulator CtrA activates transcription of roughly 100 genes, including pilA which codes for the pilin monomer that makes up the pilus filament. While most CtrA-activated promoters have a single CtrA-binding site at the - 35 position and are induced at the early to mid-predivisional cell stage, the pilA promoter has 3 additional upstream CtrA-binding sites and it is induced at the late predivisional cell stage. Reporter constructs where these additional sites were disrupted by deletion or mutation led to increased activity compared to the WT promoter. In synchronized cultures, these mutations caused pilA transcription to occur approximately 20 min earlier than WT. The results suggested that the site overlapping the - 35 position drives pilA gene expression while the other upstream CtrA-binding sites serve to reduce and delay expression. EMSA experiments showed that the - 35 Site has lower affinity for CtrA∼P compared to the other sites, suggesting binding site affinity may be involved in the delay mechanism. Mutating the upstream inhibitory CtrA-binding sites in the pilA promoter caused significantly higher numbers of pre-divisional cells to express pili, and phage survival assays showed this strain to be significantly more sensitive to pilitropic phage. These results suggest that pilA regulation evolved in C. crescentus to provide an ecological advantage within the context of phage infection.
Keywords: Caulobacter crescentus; pilA; CtrA; Transcriptional regulation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

