N6-methyladenosine writer METTL16-mediated alternative splicing and translation control are essential for murine spermatogenesis
- PMID: 39030605
- PMCID: PMC11264951
- DOI: 10.1186/s13059-024-03332-5
N6-methyladenosine writer METTL16-mediated alternative splicing and translation control are essential for murine spermatogenesis
Abstract
Background: The mitosis-to-meiosis switch during spermatogenesis requires dynamic changes in gene expression. However, the regulation of meiotic transcriptional and post-transcriptional machinery during this transition remains elusive.
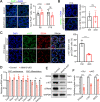
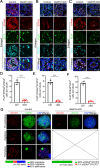
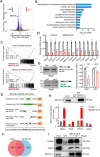
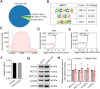
Results: We report that methyltransferase-like protein 16 (METTL16), an N6-methyladenosine (m6A) writer, is required for mitosis-to-meiosis transition during spermatogenesis. Germline conditional knockout of Mettl16 in male mice impairs spermatogonial differentiation and meiosis initiation. Mechanistically, METTL16 interacts with splicing factors to regulate the alternative splicing of meiosis-related genes such as Stag3. Ribosome profiling reveals that the translation efficiency of many meiotic genes is dysregulated in METTL16-deficient testes. m6A-sequencing shows that ablation of METTL16 causes upregulation of the m6A-enriched transcripts and downregulation of the m6A-depleted transcripts, similar to Meioc and/or Ythdc2 mutants. Further in vivo and in vitro experiments demonstrate that the methyltransferase activity site (PP185-186AA) of METTL16 is necessary for spermatogenesis.
Conclusions: Our findings support a molecular model wherein the m6A writer METTL16-mediated alternative splicing and translation efficiency regulation are required to control the mitosis-to-meiosis germ cell fate decision in mice, with implications for understanding meiosis-related male fertility disorders.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

