A chromosome-level genome assembly of Drosophila madeirensis, a fruit fly species endemic to the island of Madeira
- PMID: 39031588
- PMCID: PMC11373663
- DOI: 10.1093/g3journal/jkae167
A chromosome-level genome assembly of Drosophila madeirensis, a fruit fly species endemic to the island of Madeira
Abstract
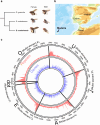
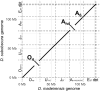
Drosophila subobscura is distributed across Europe, the Near East, and the Americas, while its sister species, Drosophila madeirensis, is endemic to the island of Madeira in the Atlantic Ocean. D. subobscura is known for its strict light-dependence in mating and its unique courtship displays, including nuptial gift-giving. D. subobscura has also attracted the interest of researchers because of its abundant variations in chromosomal polymorphisms correlated to the latitude and season, which have been used as a tool to track global climate warming. Although D. madeirensis can be an important resource for understanding the evolutionary underpinning of these genetic characteristics of D. subobscura, little work has been done on the biology of this species. Here, we used a HiFi long-read sequencing data set to produce a de novo genome assembly for D. madeirensis. This assembly comprises a total of 111 contigs spanning 135.5 Mb and has an N50 of 24.2 Mb and a BUSCO completeness score of 98.6%. Each of the 6 chromosomes of D. madeirensis consisted of a single contig except for some centromeric regions. Breakpoints of the chromosomal inversions between D. subobscura and D. madeirensis were characterized using this genome assembly, updating some of the previously identified locations.
Keywords: Drosophila madeirensis; HiFi; genome assembly; inversion breakpoint.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflict of interest The author(s) declare that they have no conflict of interest.
Figures

References
-
- Ayala FJ, Serra L, Prevosti A. 1989. A grand experiment in evolution: the Drosophila subobscura colonization of the Americas. Genome. 31(1):246–255. doi: 10.1139/g89-042. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
