Novel chimeric antigen receptors for the effective and safe treatment of NY-BR-1 positive breast cancer
- PMID: 39032146
- PMCID: PMC11260171
- DOI: 10.1002/ctm2.1776
Novel chimeric antigen receptors for the effective and safe treatment of NY-BR-1 positive breast cancer
Conflict of interest statement
Dirk Jäger, Inka Zörnig and Patrick Schmidt are owners and inventors on the patents WO2023/083982 and WO2023/083985 regarding the commercial use of CAR‐T cells targeting NY‐BR‐1.
Figures
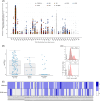

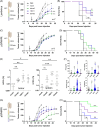

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
