Benzoxazole-derivatives enhance progranulin expression and reverse the aberrant lysosomal proteome caused by GRN haploinsufficiency
- PMID: 39033178
- PMCID: PMC11271458
- DOI: 10.1038/s41467-024-50076-8
Benzoxazole-derivatives enhance progranulin expression and reverse the aberrant lysosomal proteome caused by GRN haploinsufficiency
Abstract
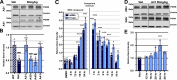
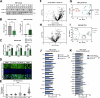
Heterozygous loss-of-function mutations in the GRN gene are a major cause of hereditary frontotemporal dementia. The mechanisms linking frontotemporal dementia pathogenesis to progranulin deficiency are not well understood, and there is currently no treatment. Our strategy to prevent the onset and progression of frontotemporal dementia in patients with GRN mutations is to utilize small molecule positive regulators of GRN expression to boost progranulin levels from the remaining functional GRN allele, thus restoring progranulin levels back to normal within the brain. This work describes a series of blood-brain-barrier-penetrant small molecules which significantly increase progranulin protein levels in human cellular models, correct progranulin protein deficiency in Grn+/- mouse brains, and reverse lysosomal proteome aberrations, a phenotypic hallmark of frontotemporal dementia, more efficiently than the previously described small molecule suberoylanilide hydroxamic acid. These molecules will allow further elucidation of the cellular functions of progranulin and its role in frontotemporal dementia and will also serve as lead structures for further drug development.
© 2024. The Author(s).
Conflict of interest statement
J.H. is a co-founder of Reelin Therapeutics Inc. and coinventor of a patent related to anti-Reelin strategies (Application Number: 15/763,047 and Publication Number: 20180273637). The remaining authors declare no competing interest.
Figures




References
MeSH terms
Substances
Grants and funding
- R01 NS093382/NS/NINDS NIH HHS/United States
- NS108115,/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- R01 HL063762/HL/NHLBI NIH HHS/United States
- R01 NS108115/NS/NINDS NIH HHS/United States
- RF1 AG053391/AG/NIA NIH HHS/United States
- P30 CA142543/CA/NCI NIH HHS/United States
- A20135245/BrightFocus Foundation (BrightFocus)
- NS093382,/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- R37 HL063762/HL/NHLBI NIH HHS/United States
- A2016396S/BrightFocus Foundation (BrightFocus)
- AG053391/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
LinkOut - more resources
Full Text Sources
Miscellaneous

