Small duct and large duct type intrahepatic cholangiocarcinoma reveal distinct patterns of immune signatures
- PMID: 39034327
- PMCID: PMC11271402
- DOI: 10.1007/s00432-024-05888-y
Small duct and large duct type intrahepatic cholangiocarcinoma reveal distinct patterns of immune signatures
Abstract
Purpose: Dedicated gene signatures in small (SD-iCCA) and large (LD-iCCA) duct type intrahepatic cholangiocarcinoma remain unknown. We performed immune profiling in SD- and LD-iCCA to identify novel biomarker candidates for personalized medicine.
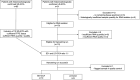
Methods: Retrospectively, 19 iCCA patients with either SD-iCCA (n = 10, median age, 63.1 years (45-86); men, 4) or LD-iCCA (n = 9, median age, 69.7 years (62-85); men, 5)) were included. All patients were diagnosed and histologically confirmed between 04/2009 and 01/2021. Tumor tissue samples were processed for differential expression profiling using NanoString nCounter® PanCancer Immune Profiling Panel.
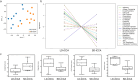
Results: With the exception of complement signatures, immune-related pathways were broadly downregulated in SD-iCCA vs. LD-iCCA. A total of 20 immune-related genes were strongly downregulated in SD-iCCA with DMBT1 (log2fc = -5.39, p = 0.01) and CEACAM6 (log2fc = -6.38, p = 0.01) showing the strongest downregulation. Among 7 strongly (log2fc > 2, p ≤ 0.02) upregulated genes, CRP (log2fc = 5.06, p = 0.02) ranked first, and four others were associated with complement (C5, C4BPA, C8A, C8B). Total tumor-infiltrating lymphocytes (TIL) signature was decreased in SD-iCCA with elevated ratios of exhausted-CD8/TILs, NK/TILs, and cytotoxic cells/TILs while having decreased ratios of B-cells/TILs, mast cells/TILs and dendritic cells/TILs. The immune profiling signatures in SD-iCCA revealed downregulation in chemokine signaling pathways inclulding JAK2/3 and ERK1/2 as well as nearly all cytokine-cytokine receptor interaction pathways with the exception of the CXCL1/CXCR1-axis.
Conclusion: Immune patterns differed in SD-iCCA versus LD-iCCA. We identified potential biomarker candidate genes, including CRP, CEACAM6, DMBT1, and various complement factors that could be explored for augmented diagnostics and treatment decision-making.
Keywords: Biomarker; Cholangiocarcinoma; Immune profiling; Intrahepatic; Pathology, molecular; Surgical oncology.
© 2024. The Author(s).
Conflict of interest statement
F. F. has received travel support from Ipsen, Abbvie, Astrazeneca and speaker’s fees from AbbVie, MSD, Ipsen, Astra. S.Z. has received speaker fees and/or honoraria for consultancy from Abbvie, BioMarin, Gilead, Intercept, Janssen, Madrigal, MSD/Merck, NovoNordisk, SoBi and Theratechnologies. P.J.W. has received consulting fees and honoraria for lectures by Bayer, Janssen-Cilag, Novartis, Roche, MSD, Astellas Pharma, Bristol-Myers Squibb, Thermo Fisher Scientific, Molecular Health, Guardant Health, Sophia Genetics, Qiagen, Eli Lilly, Myriad, Hedera Dx, and Astra Zeneca; research support was provided by Astra Zeneca. The authors declare that there is no relationship relevant to the manuscripts’ subject. All other authors declare no conflicts of interest.
Figures


References
-
- Aishima S, Oda Y (2015) Pathogenesis and classification of intrahepatic cholangiocarcinoma: different characters of perihilar large duct type versus peripheral small duct type. J Hepatobiliary Pancreat Sci 22:94–100 - PubMed
-
- Banales JM, Marin JJG, Lamarca A, Rodrigues PM, Khan SA, Roberts LR, Cardinale V, Carpino G, Andersen JB, Braconi C, Calvisi DF, Perugorria MJ, Fabris L, Boulter L, Macias RIR, Gaudio E, Alvaro D, Gradilone SA, Strazzabosco M, Marzioni M, Coulouarn C, Fouassier L, Raggi C, Invernizzi P, Mertens JC, Moncsek A, Rizvi S, Heimbach J, Koerkamp BG, Bruix J, Forner A, Bridgewater J, Valle JW, Gores GJ (2020) Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat Rev Gastroenterol Hepatol 17:557–588 - PMC - PubMed
-
- Bathum Nexoe A, Pedersen AA, von Huth S, Detlefsen S, Hansen PL, Holmskov U (2020) Immunohistochemical Localization of Deleted in Malignant Brain Tumors 1 in Normal Human Tissues. J Histochem Cytochem 68:377–387 - PubMed
-
- Bertuccio P, Bosetti C, Levi F, Decarli A, Negri E, La Vecchia C (2013) A comparison of trends in mortality from primary liver cancer and intrahepatic cholangiocarcinoma in Europe. Ann Oncol 24:1667–1674 - PubMed
-
- Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI, Ostrand-Rosenberg S, Hedrick CC, Vonderheide RH, Pittet MJ, Jain RK, Zou W, Howcroft TK, Woodhouse EC, Weinberg RA, Krummel MF (2018) Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med 24:541–550 - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

