STAT3 in acute myeloid leukemia facilitates natural killer cell-mediated surveillance
- PMID: 39034990
- PMCID: PMC11257888
- DOI: 10.3389/fimmu.2024.1374068
STAT3 in acute myeloid leukemia facilitates natural killer cell-mediated surveillance
Abstract
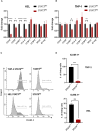
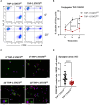
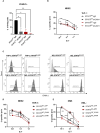
Acute myeloid leukemia (AML) is a heterogenous disease characterized by the clonal expansion of myeloid progenitor cells. Despite recent advancements in the treatment of AML, relapse still remains a significant challenge, necessitating the development of innovative therapies to eliminate minimal residual disease. One promising approach to address these unmet clinical needs is natural killer (NK) cell immunotherapy. To implement such treatments effectively, it is vital to comprehend how AML cells escape the NK-cell surveillance. Signal transducer and activator of transcription 3 (STAT3), a component of the Janus kinase (JAK)-STAT signaling pathway, is well-known for its role in driving immune evasion in various cancer types. Nevertheless, the specific function of STAT3 in AML cell escape from NK cells has not been deeply investigated. In this study, we unravel a novel role of STAT3 in sensitizing AML cells to NK-cell surveillance. We demonstrate that STAT3-deficient AML cell lines are inefficiently eliminated by NK cells. Mechanistically, AML cells lacking STAT3 fail to form an immune synapse as efficiently as their wild-type counterparts due to significantly reduced surface expression of intercellular adhesion molecule 1 (ICAM-1). The impaired killing of STAT3-deficient cells can be rescued by ICAM-1 overexpression proving its central role in the observed phenotype. Importantly, analysis of our AML patient cohort revealed a positive correlation between ICAM1 and STAT3 expression suggesting a predominant role of STAT3 in ICAM-1 regulation in this disease. In line, high ICAM1 expression correlates with better survival of AML patients underscoring the translational relevance of our findings. Taken together, our data unveil a novel role of STAT3 in preventing AML cells from escaping NK-cell surveillance and highlight the STAT3/ICAM-1 axis as a potential biomarker for NK-cell therapies in AML.
Keywords: AML; ICAM-1; NK cells; STAT3; immunotherapy.
Copyright © 2024 Witalisz-Siepracka, Denk, Zdársky, Hofmann, Edtmayer, Harm, Weiss, Heindl, Hessenberger, Summer, Dutta, Casanova, Obermair, Győrffy, Putz, Sill and Stoiber.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Pelcovits A, Niroula R. Acute myeloid leukemia: A review. R I Med J (2013). (2020) 103:38–40. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

