Complex SMN Hybrids Detected in a Cohort of 31 Patients With Spinal Muscular Atrophy
- PMID: 39035824
- PMCID: PMC11259531
- DOI: 10.1212/NXG.0000000000200175
Complex SMN Hybrids Detected in a Cohort of 31 Patients With Spinal Muscular Atrophy
Erratum in
-
Erratum: Complex SMN Hybrids Detected in a Cohort of 31 Patients With Spinal Muscular Atrophy.Neurol Genet. 2024 Oct 23;10(6):e200212. doi: 10.1212/NXG.0000000000200212. eCollection 2024 Dec. Neurol Genet. 2024. PMID: 39807214 Free PMC article.
Abstract
Background and objectives: Spinal muscular atrophy (SMA) is a recessive neuromuscular disorder caused by the loss or presence of point pathogenic variants in the SMN1 gene. The main positive modifier of the SMA phenotype is the number of copies of the SMN2 gene, a paralog of SMN1, which only produces around 10%-15% of functional SMN protein. The SMN2 copy number is inversely correlated with phenotype severity; however, discrepancies between the SMA type and the SMN2 copy number have been reported. The presence of SMN2-SMN1 hybrids has been proposed as a possible modifier of SMA disease.
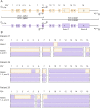
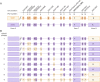
Methods: We studied 31 patients with SMA, followed at a single center and molecularly diagnosed by Multiplex Ligand-Dependent Probe Amplification (MLPA), with a specific next-generation sequencing protocol to investigate their SMN2 genes in depth. Hybrid characterization also included bioinformatics haplotype phasing and specific PCRs to resolve each SMN2-SMN1 hybrid structure.
Results: We detected SMN2-SMN1 hybrid genes in 45.2% of the patients (14/31), the highest rate reported to date. This represents a total of 25 hybrid alleles, with 9 different structures, of which only 4 are detectable by MLPA. Of particular interest were 2 patients who presented 4 SMN2-SMN1 hybrid copies each and no pure SMN2 copies, an event reported here for the first time. No clear trend between the presence of hybrids and a milder phenotype was observed, although 5 of the patients with hybrid copies showed a better-than-expected phenotype. The higher hybrid detection rate in our cohort may be due to both the methodology applied, which allows an in-depth characterization of the SMN genes and the ethnicity of the patients, mainly of African origin.
Discussion: Although hybrid genes have been proposed to be beneficial for patients with SMA, our work revealed great complexity and variability between hybrid structures; therefore, each hybrid structure should be studied independently to determine its contribution to the SMA phenotype. Large-scale studies are needed to gain a better understanding of the function and implications of SMN2-SMN1 hybrid copies, improving genotype-phenotype correlations and prediction of the evolution of patients with SMA.
Copyright © 2024 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of the American Academy of Neurology.
Conflict of interest statement
E.F. Tizzano has served as a consultant and has participated on advisory boards for Novartis Gene Therapies, Inc., Biogen, Biologix, Cytokinetics, Novartis, and Roche, and research funding from Biogen/Ionis and Roche. S. Quijano-Roy is a site principal investigator for clinical trials of Biogen and Novartis Gene Therapies, Inc.; has served as a consultant and has participated on advisory boards for Novartis Gene Therapies, Inc., Biogen, and Roche; and has received travel and speaker honoraria from Biogen, Novartis, and Roche. Go to Neurology.org/NG for full disclosures.
Figures

References
LinkOut - more resources
Full Text Sources
