Genetic characterization and estimated 4CMenB vaccine strain coverage of 284 Neisseria meningitidis isolates causing invasive meningococcal disease in Argentina in 2010-2014
- PMID: 39037011
- PMCID: PMC11789736
- DOI: 10.1080/21645515.2024.2378537
Genetic characterization and estimated 4CMenB vaccine strain coverage of 284 Neisseria meningitidis isolates causing invasive meningococcal disease in Argentina in 2010-2014
Abstract
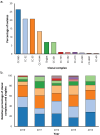
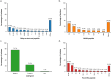
Meningococcal (Neisseria meningitidis) serogroup B (MenB) strain antigens are diverse and a limited number of strains can be evaluated using the human serum bactericidal antibody (hSBA) assay. The genetic Meningococcal Antigen Typing System (gMATS) was developed to predict the likelihood of coverage for large numbers of isolates by the 4CMenB vaccine, which includes antigens Neisseria adhesin A (NadA), Neisserial Heparin-Binding Antigen (NHBA), factor H-binding protein (fHbp), and Porin A (PorA). In this study, we characterized by whole-genome analyses 284 invasive MenB isolates collected from 2010 to 2014 by the Argentinian National Laboratories Network (52-61 isolates per year). Strain coverage was estimated by gMATS on all isolates and by hSBA assay on 74 randomly selected isolates, representative of the whole panel. The four most common clonal complexes (CCs), accounting for 81.3% of isolates, were CC-865 (75 isolates, 26.4%), CC-32 (59, 20.8%), CC-35 (59, 20.8%), and CC-41/44 (38, 13.4%). Vaccine antigen genotyping showed diversity. The most prevalent variants/peptides were fHbp variant 2, NHBA peptides 24, 21, and 2, and PorA variable region 2 profiles 16-36 and 14. The nadA gene was present in 66 (23.2%) isolates. Estimated strain coverage by hSBA assay showed 78.4% of isolates were killed by pooled adolescent sera, and 51.4% and 64.9% (based on two different thresholds) were killed by pooled infant sera. Estimated coverage by gMATS (61.3%; prediction interval: 55.5%, 66.7%) was consistent with the infant hSBA assay results. Continued genomic surveillance is needed to evaluate the persistence of major MenB CCs in Argentina.
Keywords: 4CMenB; Argentina; Neisseria meningitidis; gMATS; human serum bactericidal assay; meningococcal disease; serogroup B; vaccine; whole genome sequencing.
Plain language summary
The most common clinical manifestations of invasive meningococcal disease include meningitis and septicemia, which can be deadly, and many survivors suffer long-term serious after-effects. Most cases of invasive meningococcal disease are caused by six meningococcal serogroups (types), including serogroup B. Although vaccines are available against meningococcal serogroup B infection, these vaccines target antigens that are highly diverse. Consequently, the effectiveness of vaccination may vary from country to country because the meningococcal serogroup B strains circulating in particular regions carry different forms of the target vaccine antigens. This means it is important to test serogroup B strains isolated from specific populations to estimate the percentage of strains that a vaccine is likely to be effective against (known as ‘vaccine strain coverage’). The genetic Meningococcal Antigen Typing System (gMATS) was developed to predict strain coverage by the four-component meningococcal serogroup B vaccine, 4CMenB, against large numbers of serogroup B strains. In this study, we analyzed 284 invasive meningococcal serogroup B isolates collected between 2010 and 2014 in Argentina. Genetic analyses showed that the vaccine antigens of the isolates were diverse and some genetic characteristics had not been found in isolates from other countries. However, vaccine strain coverage estimated by gMATS was consistent with that reported in other parts of the world and with strain coverage results obtained for a subset via another method, the human serum bactericidal antibody (hSBA) assay. These results highlight the need for continued monitoring of circulating bacterial strains to assess the estimated strain coverage of meningococcal serogroup B vaccines.
Conflict of interest statement
Alessandro Brozzi, Alessia Biolchi, Margherita Bodini, Maria Giuliani, Silvia Guidotti, Alessandro Muzzi, Florencia Nocita, and Sara Tomei are employed by GSK. Mariagrazia Pizza, Rino Rappuoli, and Gabriela Vidal were employed by GSK at the time of the study. Alessia Biolchi and Alessandro Muzzi hold financial equities in GSK. Carla Vizzotti was EPI manager of the Ministry of Health in Argentina from 2010 to 2015, Vice-Minister of Health from 2019 to 2021, and Minister of Health from 2021 to 2023. Josefina Campos was employed by ANLIS “Dr. Carlos G. Malbrán” at the time of the study. She also received grants from the Pan American Health Organization, Wellcome Trust, and EMBL European Bioinformatics Institute; payment/honoraria for lectures from the Wellcome Trust Sanger Institute; and support for attending meetings and/or travel from the Pan American Health Organization, the World Health Organization, and the European Society in Clinical Microbiology and Infectious Diseases. These authors declare no other financial and non-financial relationships and activities. Adriana Efron, Federico Lorenzo, María Alicia Moscoloni, and Cecilia Sorhouet Pereira declare no financial and non-financial relationships and activities and no conflicts of interest.
Figures

References
-
- Parikh S, Campbell H, Bettinger JA, Harrison LH, Marshall HS, Martinon-Torres F, Safadi MA, Shao Z, Zhu B, von Gottberg A, et al. The everchanging epidemiology of meningococcal disease worldwide and the potential for prevention through vaccination. J Infect. 2020;81(4):483–98. doi:10.1016/j.jinf.2020.05.079. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
