SARS-CoV-2 infection predisposes patients to coinfection with Staphylococcus aureus
- PMID: 39037272
- PMCID: PMC11323729
- DOI: 10.1128/mbio.01667-24
SARS-CoV-2 infection predisposes patients to coinfection with Staphylococcus aureus
Abstract
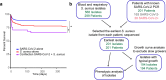
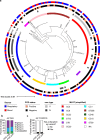
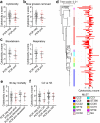
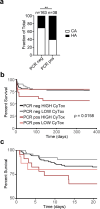
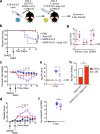
Severe COVID-19 has been associated with coinfections with bacterial and fungal pathogens. Notably, patients with COVID-19 who develop Staphylococcus aureus bacteremia exhibit higher rates of mortality than those infected with either pathogen alone. To understand this clinical scenario, we collected and examined S. aureus blood and respiratory isolates from a hospital in New York City during the early phase of the pandemic from both SARS-CoV-2+ and SARS-CoV-2- patients. Whole genome sequencing of these S. aureus isolates revealed broad phylogenetic diversity in both patient groups, suggesting that SARS-CoV-2 coinfection was not associated with a particular S. aureus lineage. Phenotypic characterization of the contemporary collection of S. aureus isolates from SARS-CoV-2+ and SARS-CoV-2- patients revealed no notable differences in several virulence traits examined. However, we noted a trend toward overrepresentation of S. aureus bloodstream strains with low cytotoxicity in the SARS-CoV-2+ group. We observed that patients coinfected with SARS-CoV-2 and S. aureus were more likely to die during the acute phase of infection when the coinfecting S. aureus strain exhibited high or low cytotoxicity. To further investigate the relationship between SARS-CoV-2 and S. aureus infections, we developed a murine coinfection model. These studies revealed that infection with SARS-CoV-2 renders mice susceptible to subsequent superinfection with low cytotoxicity S. aureus. Thus, SARS-CoV-2 infection sensitizes the host to coinfections, including S. aureus isolates with low intrinsic virulence.
Importance: The COVID-19 pandemic has had an enormous impact on healthcare across the globe. Patients who were severely infected with SARS-CoV-2, the virus causing COVID-19, sometimes became infected with other pathogens, which is termed coinfection. If the coinfecting pathogen is the bacterium Staphylococcus aureus, there is an increased risk of patient death. We collected S. aureus strains that coinfected patients with SARS-CoV-2 to study the disease outcome caused by the interaction of these two important pathogens. We found that both in patients and in mice, coinfection with an S. aureus strain lacking toxicity resulted in more severe disease during the early phase of infection, compared with infection with either pathogen alone. Thus, SARS-CoV-2 infection can directly increase the severity of S. aureus infection.
Keywords: COVID; MRSA; SARS-CoV-2; agr; coinfection.
Conflict of interest statement
V.J.T. has consulted for Janssen Research & Development, LLC, and has received honoraria from Genentech and Medimmune. He is also an inventor on patents and patent applications filed by New York University, which are currently under commercial license to Janssen Biotech Inc. Janssen Biotech Inc. had provided research funding and other payments associated with a licensing agreement. B.S. has consulted for Regeneron and MicroGenDx. K.C. has received research support from Pfizer, Takeda, Pacific Biosciences, Genentech, and AbbVie. R.T. completed a NYU-Regeneron postdoctoral training program in laboratory animal medicine.
Figures
References
-
- Jakab GJ. 1981. Mechanisms of virus-induced bacterial superinfections of the lung. Clin Chest Med 2:59–66. - PubMed
MeSH terms
Grants and funding
- AI143639/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- AI137336/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01 AI099394/AI/NIAID NIH HHS/United States
- R01 AI140754/AI/NIAID NIH HHS/United States
- R01 AI105129/AI/NIAID NIH HHS/United States
- AI105129/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01 AI137336/AI/NIAID NIH HHS/United States
- AI099394/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- AI140754/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01 AI143639/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

