Cognitive rejuvenation in old rats by hippocampal OSKM gene therapy
- PMID: 39037528
- PMCID: PMC11872836
- DOI: 10.1007/s11357-024-01269-y
Cognitive rejuvenation in old rats by hippocampal OSKM gene therapy
Abstract
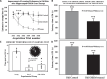
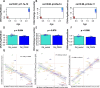
Several studies have indicated that interrupted epigenetic reprogramming using Yamanaka transcription factors (OSKM) can rejuvenate cells from old laboratory animals and humans. However, the potential of OSKM-induced rejuvenation in brain tissue has been less explored. Here, we aimed to restore cognitive performance in 25.3-month-old female Sprague-Dawley rats using OSKM gene therapy for 39 days. Their progress was then compared with the cognitive performance of untreated 3.5-month-old rats as well as old control rats treated with a placebo adenovector. The Barnes maze test, used to assess cognitive performance, demonstrated enhanced cognitive abilities in old rats treated with OSKM compared to old control animals. In the treated old rats, there was a noticeable trend towards improved spatial memory relative to the old controls. Further, OSKM gene expression did not lead to any pathological alterations within the 39 days. Analysis of DNA methylation following OSKM treatment yielded three insights. First, epigenetic clocks for rats suggested a marginally significant epigenetic rejuvenation. Second, chromatin state analysis revealed that OSKM treatment rejuvenated the methylome of the hippocampus. Third, an epigenome-wide association analysis indicated that OSKM expression in the hippocampus of old rats partially reversed the age-related increase in methylation. In summary, the administration of Yamanaka genes via viral vectors rejuvenates the functional capabilities and the epigenetic landscape of the rat hippocampus.
Keywords: Epigenetic age; Hippocampal aging; OSKM gene therapy; OSKM-induced demethylation; Rejuvenation; Spatial memory.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interest: Steve Horvath is a founder of the non-profit Epigenetic Clock Development Foundation which licenses several patents surrounding methylation measurements and epigenetic clock testing from his former employer UC Regents. These patents list SH as inventor. Robert T. Brooke is a founder and CEO of the Clock foundation. SH is an employee of Altos Labs Inc, UK. The other authors declare no conflicts of interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

