Recombinant origin and interspecies transmission of a HERV-K(HML-2)-related primate retrovirus with a novel RNA transport element
- PMID: 39037763
- PMCID: PMC11379458
- DOI: 10.7554/eLife.80216
Recombinant origin and interspecies transmission of a HERV-K(HML-2)-related primate retrovirus with a novel RNA transport element
Abstract
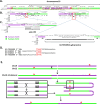
HERV-K(HML-2), the youngest clade of human endogenous retroviruses (HERVs), includes many intact or nearly intact proviruses, but no replication competent HML-2 proviruses have been identified in humans. HML-2-related proviruses are present in other primates, including rhesus macaques, but the extent and timing of HML-2 activity in macaques remains unclear. We have identified 145 HML-2-like proviruses in rhesus macaques, including a clade of young, rhesus-specific insertions. Age estimates, intact open reading frames, and insertional polymorphism of these insertions are consistent with recent or ongoing infectious activity in macaques. 106 of the proviruses form a clade characterized by an ~750 bp sequence between env and the 3' long terminal repeat (LTR), derived from an ancient recombination with a HERV-K(HML-8)-related virus. This clade is found in Old World monkeys (OWM), but not great apes, suggesting it originated after the ape/OWM split. We identified similar proviruses in white-cheeked gibbons; the gibbon insertions cluster within the OWM recombinant clade, suggesting interspecies transmission from OWM to gibbons. The LTRs of the youngest proviruses have deletions in U3, which disrupt the Rec Response Element (RcRE), required for nuclear export of unspliced viral RNA. We show that the HML-8-derived region functions as a Rec-independent constitutive transport element (CTE), indicating the ancestral Rec-RcRE export system was replaced by a CTE mechanism.
Keywords: RNA export; endogenous retrovirus; evolutionary biology; human; infectious disease; microbiology; recombination; rhesus macaque; viral evolution; viruses; zoonosis.
Plain language summary
Just as we study fossils to understand how animals and plants have evolved, we can study ancient viruses to understand how diseases have emerged and changed over long periods. Unlike fossils, viruses do not leave visible traces in the ground but, instead, they leave viral genes known as endogenous viral elements (or EVEs) that become permanently incorporated in their host’s DNA. HML-2s are the youngest known EVEs in the human genome. They have evolved gradually by accumulating lots of small genetic changes and no longer actively infect humans. But these virus remnants have long been suspected to play a role in prostate cancer, lupus and other human diseases. Rhesus macaques and other monkeys also have HML-2s but these are less well studied than human HML-2s. Monkeys are often used as models of human biology in research studies, therefore, understanding how HML-2s have evolved in rhesus macaques may enable researchers to establish this monkey as a model for investigating the role of HML-2s in humans. To investigate this possibility, Williams et al. searched for HML-like EVEs in rhesus macaque genomes published in previous studies. The experiments found that, unlike human HML-2s, the macaque HML-2s underwent a sudden genetic transformation millions of years ago. They acquired a new gene from another virus that completely changed how the macaque HML-2s leave a compartment within the cells of their host that contains most of the host’s genome – a key step in the life cycle of viruses. The data also suggest that HML-2s may still be actively infecting macaques today and that these EVEs jumped from monkeys into gibbons. This is the first known example of HML-2s moving between different types of primates and it indicates there may be a risk that macaque HML-2s could infect humans. In the future, the findings of Williams et al. may help researchers develop new approaches to treat prostate cancer and other diseases linked with HML-2s in humans.
© 2024, Williams et al.
Conflict of interest statement
ZW, AI, LG, DL, SM, JP, JC, WJ No competing interests declared
Figures













Update of
- doi: 10.1101/2022.05.11.490678
References
-
- Bamunusinghe D, Liu Q, Plishka R, Dolan MA, Skorski M, Oler AJ, Yedavalli VRK, Buckler-White A, Hartley JW, Kozak CA. Recombinant origins of pathogenic and nonpathogenic mouse gammaretroviruses with polytropic host range. Journal of Virology. 2017;91:e00855-17. doi: 10.1128/JVI.00855-17. - DOI - PMC - PubMed
-
- Bergeron LA, Besenbacher S, Bakker J, Zheng J, Li P, Pacheco G, Sinding MHS, Kamilari M, Gilbert MTP, Schierup MH, Zhang G. The germline mutational process in rhesus macaque and its implications for phylogenetic dating. GigaScience. 2021;10:giab029. doi: 10.1093/gigascience/giab029. - DOI - PMC - PubMed
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

