Prochlorococcus marinus responses to light and oxygen
- PMID: 39038009
- PMCID: PMC11262661
- DOI: 10.1371/journal.pone.0307549
Prochlorococcus marinus responses to light and oxygen
Abstract
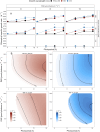
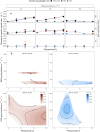
Prochlorococcus marinus, the smallest picocyanobacterium, comprises multiple clades occupying distinct niches, currently across tropical and sub-tropical oligotrophic ocean regions, including Oxygen Minimum Zones. Ocean warming may open growth-permissive temperatures in new, poleward photic regimes, along with expanded Oxygen Minimum Zones. We used ocean metaproteomic data on current Prochlorococcus marinus niches, to guide testing of Prochlorococcus marinus growth across a matrix of peak irradiances, photoperiods, spectral bands and dissolved oxygen. MED4 from Clade HLI requires greater than 4 h photoperiod, grows at 25 μmol O2 L-1 and above, and exploits high cumulative diel photon doses. MED4, however, relies upon an alternative oxidase to balance electron transport, which may exclude it from growth under our lowest, 2.5 μmol O2 L-1, condition. SS120 from clade LLII/III is restricted to low light under full 250 μmol O2 L-1, shows expanded light exploitation under 25 μmol O2 L-1, but is excluded from growth under 2.5 μmol O2 L-1. Intermediate oxygen suppresses the cost of PSII photoinactivation, and possibly the enzymatic production of H2O2 in SS120, which has limitations on genomic capacity for PSII and DNA repair. MIT9313 from Clade LLIV is restricted to low blue irradiance under 250 μmol O2 L-1, but exploits much higher irradiance under red light, or under lower O2 concentrations, conditions which slow photoinactivation of PSII and production of reactive oxygen species. In warming oceans, range expansions and competition among clades will be governed not only by light levels. Short photoperiods governed by latitude, temperate winters, and depth attenuation of light, will exclude clade HLI (including MED4) from some habitats. In contrast, clade LLII/III (including SS120), and particularly clade LLIV (including MIT9313), may exploit higher light niches nearer the surface, under expanding OMZ conditions, where low O2 relieves the stresses of oxidation stress and PSII photoinhibition.
Copyright: © 2024 Savoie et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures








Similar articles
-
Comparison of photosynthetic performances of marine picocyanobacteria with different configurations of the oxygen-evolving complex.Photosynth Res. 2018 Oct;138(1):57-71. doi: 10.1007/s11120-018-0539-3. Epub 2018 Jun 25. Photosynth Res. 2018. PMID: 29938315
-
Membrane organization of photosystem I complexes in the most abundant phototroph on Earth.Nat Plants. 2019 Aug;5(8):879-889. doi: 10.1038/s41477-019-0475-z. Epub 2019 Jul 22. Nat Plants. 2019. PMID: 31332310 Free PMC article.
-
Ultraviolet stress delays chromosome replication in light/dark synchronized cells of the marine cyanobacterium Prochlorococcus marinus PCC9511.BMC Microbiol. 2010 Jul 29;10:204. doi: 10.1186/1471-2180-10-204. BMC Microbiol. 2010. PMID: 20670397 Free PMC article.
-
A minimum set of regulators to thrive in the ocean.FEMS Microbiol Rev. 2020 Mar 1;44(2):232-252. doi: 10.1093/femsre/fuaa005. FEMS Microbiol Rev. 2020. PMID: 32077939 Review.
-
Phycobiliproteins in Prochlorococcus marinus: biosynthesis of pigments and their assembly into proteins.Eur J Cell Biol. 2010 Dec;89(12):1005-10. doi: 10.1016/j.ejcb.2010.06.017. Epub 2010 Aug 17. Eur J Cell Biol. 2010. PMID: 20724022 Review.
Cited by
-
Mesoscale eddies shape Prochlorococcus community structure and dynamics in the oligotrophic open ocean.ISME J. 2025 Jan 2;19(1):wraf106. doi: 10.1093/ismejo/wraf106. ISME J. 2025. PMID: 40415184 Free PMC article.
References
-
- Chisholm SW, Frankel SL, Goericke R, Olson RJ, Palenik B, Waterbury JB, et al. Prochlorococcus Marinus nov. Gen. Nov. Sp.: An oxyphototrophic marine prokaryote containing divinyl chlorophyll a and b. Archives of Microbiology. 1992;157: 297–300. doi: 10.1007/BF00245165 - DOI
-
- Moore LR, Goericke R, Chisholm SW. Comparative physiology of Synechococcus and Prochlorococcus: Influence of light and temperature on growth, pigments, fluorescence and absorptive properties. Marine Ecology Progress Series. 1995;116: 259–275. Available: https://www.jstor.org/stable/44635011.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

