Inducible pluripotent stem cells to study human mast cell trajectories
- PMID: 39038754
- PMCID: PMC11801248
- DOI: 10.1016/j.mucimm.2024.07.003
Inducible pluripotent stem cells to study human mast cell trajectories
Abstract
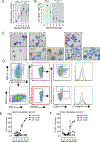
Mast cells (MCs) are derived from CD34+ hematopoietic progenitors, consist of different subtypes, and are involved in several inflammatory conditions. However, our understanding of human MC developmental trajectories and subtypes has been limited by a scarcity of suitable cellular model systems. Herein, we developed an in vitro model of human MC differentiation from induced pluripotent stem cells (iPSC) to study human MC differentiation trajectories. Flow cytometry characterization of hemopoietic cells derived from the myeloid cells-forming complex (MCFC) revealed an initial increase in Lin- CD34+ hematopoietic progenitors within Weeks 1-3, followed by an increase in CD34- CD45RA- SSClow and SSChigh hematopoietic cells. The Lin- CD34+ hematopoietic progenitors consisted of SSClow CD45RA- CD123± c-Kit+ FcεRI+ populations that were β7-integrinhigh CD203c+ and β7-integrinhigh CD203c- cells consistent with CMPFcεRI+ cells. Flow cytometry and cytologic analyses of the CD34- Lin- (SSClow) population revealed hypogranular cell populations, predominantly characterized by CD45RA- CD123± c-Kit+ FcεRI- β7-integrinlow and CD45RA- CD123± c-Kit- FcεRI+ β7-integrinMid cells. Analyses of hypergranular SSChigh cells identified Lin- CD34- CD45RA- c-Kit+ FcεRI- and Lin- CD34- CD45RA- c-Kit+ FcεRI+ cells. scRNA-seq analysis of the cells harvested at week 4 of the MCFC culture revealed the presence of monocyte and granulocyte progenitors (n = 547 cells, 26.7 %), Erythrocyte / unknown (n = 85, 4.1 %), neutrophils / myelocytes (n = 211 cells, 10.2 %), mast cell progenitor 1 (n = 599, 29.1 %), mast cell progenitor 2 (n = 152, 7.4 %), committed mast cell precursor (n = 113, 5.5 %), and MCs (n = 353, 17.1 %). In silico analyses of the MC precursor and mature MC populations revealed transcriptionally distinct MC precursor subtype and mature MC states (CMA1+ and CMA1- subtypes). Culturing MC precursor populations in MC maturation media (mast cell media II) led to homogenous mature MC populations as evidenced by high expression of high-affinity IgE receptor, metachromatic granules, presence of MC granule proteins (Tryptase and Chymase) and activation following substance P stimulation and FcεRI crosslinking. This human iPSC-based approach generates MC precursors and phenotypically mature and functional MC populations. This system will be a useful model to generate human MC populations and broaden our understanding of MC biology and transcriptional regulation of MC differentiation trajectories.
Keywords: Induced pluripotent stem cell; Mast cell; Mast cell precursors.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

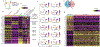



References
-
- Shah K, Ignacio A, McCoy KD, Harris NL. The emerging roles of eosinophils in mucosal homeostasis. Mucosal Immunol. 2020;13(4):574–583. - PubMed
-
- Varricchi G, Marone G, Kovanen PT. Cardiac Mast Cells: Underappreciated Immune Cells in Cardiovascular Homeostasis and Disease. Trends Immunol. 2020;41(8): 734–746. - PubMed
-
- Rothenberg ME, Hogan SP. The eosinophil. Annu Rev Immunol. 2006;24:147–174. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

