Methanogenic patterns in the gut microbiome are associated with survival in a population of feral horses
- PMID: 39039075
- PMCID: PMC11263349
- DOI: 10.1038/s41467-024-49963-x
Methanogenic patterns in the gut microbiome are associated with survival in a population of feral horses
Abstract
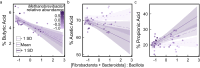
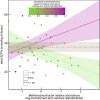
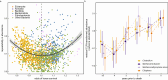
Gut microbiomes are widely hypothesised to influence host fitness and have been experimentally shown to affect host health and phenotypes under laboratory conditions. However, the extent to which they do so in free-living animal populations and the proximate mechanisms involved remain open questions. In this study, using long-term, individual-based life history and shallow shotgun metagenomic sequencing data (2394 fecal samples from 794 individuals collected between 2013-2019), we quantify relationships between gut microbiome variation and survival in a feral population of horses under natural food limitation (Sable Island, Canada), and test metagenome-derived predictions using short-chain fatty acid data. We report detailed evidence that variation in the gut microbiome is associated with a host fitness proxy in nature and outline hypotheses of pathogenesis and methanogenesis as key causal mechanisms which may underlie such patterns in feral horses, and perhaps, wild herbivores more generally.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
- 2016-06459/Canadian Network for Research and Innovation in Machining Technology, Natural Sciences and Engineering Research Council of Canada (NSERC Canadian Network for Research and Innovation in Machining Technology)
- 2019-04388/Canadian Network for Research and Innovation in Machining Technology, Natural Sciences and Engineering Research Council of Canada (NSERC Canadian Network for Research and Innovation in Machining Technology)
- 25046/Canada Foundation for Innovation (Fondation canadienne pour l'innovation)
- D20EQ-05/Morris Animal Foundation (MAF)
- D20EQ-05/Morris Animal Foundation (MAF)
LinkOut - more resources
Full Text Sources
Medical

