Magnetic-field-driven targeting of exosomes modulates immune and metabolic changes in dystrophic muscle
- PMID: 39039121
- PMCID: PMC11486659
- DOI: 10.1038/s41565-024-01725-y
Magnetic-field-driven targeting of exosomes modulates immune and metabolic changes in dystrophic muscle
Abstract
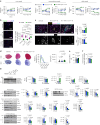
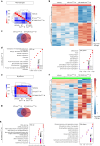
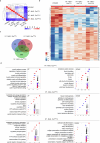
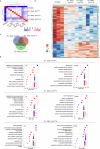
Exosomes are promising therapeutics for tissue repair and regeneration to induce and guide appropriate immune responses in dystrophic pathologies. However, manipulating exosomes to control their biodistribution and targeting them in vivo to achieve adequate therapeutic benefits still poses a major challenge. Here we overcome this limitation by developing an externally controlled delivery system for primed annexin A1 myo-exosomes (Exomyo). Effective nanocarriers are realized by immobilizing the Exomyo onto ferromagnetic nanotubes to achieve controlled delivery and localization of Exomyo to skeletal muscles by systemic injection using an external magnetic field. Quantitative muscle-level analyses revealed that macrophages dominate the uptake of Exomyo from these ferromagnetic nanotubes in vivo to synergistically promote beneficial muscle responses in a murine animal model of Duchenne muscular dystrophy. Our findings provide insights into the development of exosome-based therapies for muscle diseases and, in general, highlight the formulation of effective functional nanocarriers aimed at optimizing exosome biodistribution.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

