Macrolones target bacterial ribosomes and DNA gyrase and can evade resistance mechanisms
- PMID: 39039256
- PMCID: PMC11686707
- DOI: 10.1038/s41589-024-01685-3
Macrolones target bacterial ribosomes and DNA gyrase and can evade resistance mechanisms
Abstract
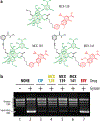
Growing resistance toward ribosome-targeting macrolide antibiotics has limited their clinical utility and urged the search for superior compounds. Macrolones are synthetic macrolide derivatives with a quinolone side chain, structurally similar to DNA topoisomerase-targeting fluoroquinolones. While macrolones show enhanced activity, their modes of action have remained unknown. Here, we present the first structures of ribosome-bound macrolones, showing that the macrolide part occupies the macrolide-binding site in the ribosomal exit tunnel, whereas the quinolone moiety establishes new interactions with the tunnel. Macrolones efficiently inhibit both the ribosome and DNA topoisomerase in vitro. However, in the cell, they target either the ribosome or DNA gyrase or concurrently both of them. In contrast to macrolide or fluoroquinolone antibiotics alone, dual-targeting macrolones are less prone to select resistant bacteria carrying target-site mutations or to activate inducible macrolide resistance genes. Furthermore, because some macrolones engage Erm-modified ribosomes, they retain activity even against strains with constitutive erm resistance genes.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
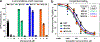


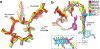




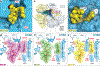


References
-
- Fernandes P Use of antibiotic core structures to generate new and useful macrolide antibiotics. in Antibiotics Current Innovations and Future Trends (eds. Sánchez S & Demain AL) (Caister Academic Press, Norfolk, UK, 2015).
-
- Agouridas C et al. Synthesis and antibacterial activity of ketolides (6-O-methyl-3-oxoerythromycin derivatives): a new class of antibacterials highly potent against macrolide-resistant and -susceptible respiratory pathogens. J. Med. Chem 41, 4080–4100 (1998). - PubMed
-
- Pavlovic D, Fajdetic A & Mutak S Novel hybrids of 15-membered 8a- and 9a-azahomoerythromycin A ketolides and quinolones as potent antibacterials. Bioorg. Med. Chem 18, 8566–8582 (2010). - PubMed
REFERENCES FOR THE ONLINE METHODS
-
- Bundy BC & Swartz JR Site-specific incorporation of p-propargyloxyphenylalanine in a cell-free environment for direct protein-protein click conjugation. Bioconjug. Chem 21, 255–63 (2010). - PubMed
-
- Cheng Y & Prusoff WH Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol 22, 3099–3108 (1973). - PubMed
MeSH terms
Substances
Grants and funding
- R35 GM127134/GM/NIGMS NIH HHS/United States
- S10 OD021527/OD/NIH HHS/United States
- R21-AI137584/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- 81673335/National Natural Science Foundation of China (National Science Foundation of China)
- P30 GM124165/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

