A common mechanism for recruiting the Rrm3 and RTEL1 accessory helicases to the eukaryotic replisome
- PMID: 39039288
- PMCID: PMC11405395
- DOI: 10.1038/s44318-024-00168-4
A common mechanism for recruiting the Rrm3 and RTEL1 accessory helicases to the eukaryotic replisome
Abstract
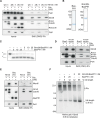
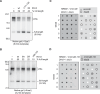
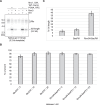
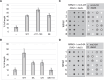
The eukaryotic replisome is assembled around the CMG (CDC45-MCM-GINS) replicative helicase, which encircles the leading-strand DNA template at replication forks. When CMG stalls during DNA replication termination, or at barriers such as DNA-protein crosslinks on the leading strand template, a second helicase is deployed on the lagging strand template to support replisome progression. How these 'accessory' helicases are targeted to the replisome to mediate barrier bypass and replication termination remains unknown. Here, by combining AlphaFold structural modelling with experimental validation, we show that the budding yeast Rrm3 accessory helicase contains two Short Linear Interaction Motifs (SLIMs) in its disordered N-terminus, which interact with CMG and the leading-strand DNA polymerase Polε on one side of the replisome. This flexible tether positions Rrm3 adjacent to the lagging strand template on which it translocates, and is critical for replication termination in vitro and Rrm3 function in vivo. The primary accessory helicase in metazoa, RTEL1, is evolutionarily unrelated to Rrm3, but binds to CMG and Polε in an analogous manner, revealing a conserved docking mechanism for accessory helicases in the eukaryotic replisome.
Keywords: Accessory Helicase; CMG Helicase; DNA Replication; RTEL1; Rrm3.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








References
-
- Bjorkman A, Johansen SL, Lin L, Schertzer M, Kanellis DC, Katsori AM, Christensen ST, Luo Y, Andersen JS, Elsasser SJ et al (2020) Human RTEL1 associates with Poldip3 to facilitate responses to replication stress and R-loop resolution. Genes Dev 34:1065–1074 10.1101/gad.330050.119 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

