Unsupervised clustering identified clinically relevant metabolic syndrome endotypes in UK and Taiwan Biobanks
- PMID: 39040048
- PMCID: PMC11260869
- DOI: 10.1016/j.isci.2024.109815
Unsupervised clustering identified clinically relevant metabolic syndrome endotypes in UK and Taiwan Biobanks
Abstract
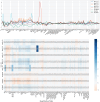
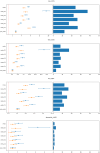
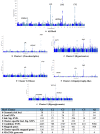
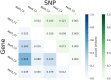
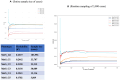
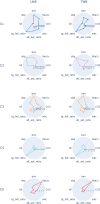
Metabolic syndrome (MetS) is a collection of cardiovascular risk factors; however, the high prevalence and heterogeneity impede effective clinical management. We conducted unsupervised clustering on individuals from UK Biobank to reveal endotypes. Five MetS subgroups were identified: Cluster 1 (C1): non-descriptive, Cluster 2 (C2): hypertensive, Cluster 3 (C3): obese, Cluster 4 (C4): lipodystrophy-like, and Cluster 5 (C5): hyperglycemic. For all of the endotypes, we identified the corresponding cardiometabolic traits and their associations with clinical outcomes. Genome-wide association studies (GWASs) were conducted to identify associated genotypic traits. We then determined endotype-specific genotypic traits and constructed polygenic risk score (PRS) models specific to each endotype. GWAS of each MetS clusters revealed different genotypic traits. C1 GWAS revealed novel findings of TRIM63, MYBPC3, MYLPF, and RAPSN. Intriguingly, C1, C3, and C4 were associated with genes highly expressed in brain tissues. MetS clusters with comparable phenotypic and genotypic traits were identified in Taiwan Biobank.
Keywords: Association analysis; Classification of bioinformatical subject; Human metabolism; Risk stratification; Syndrome.
© 2024 Institute of Biomedical Sciences, Academia Sinica, Taiwan.
Conflict of interest statement
All authors declare no competing interests.
Figures


References
-
- Duarte C.W., Vaughan L.K., Beasley T.M., Tiwari H.K. In: Emery and Rimoin's Principles and Practice of Medical Genetics. Sixth Edition. Rimoin D., Pyeritz R., Korf B., editors. Academic Press; 2013. Chapter 12 - Multifactorial Inheritance and Complex Diseases; pp. 1–15. - DOI
-
- Malik R., Chauhan G., Traylor M., Sargurupremraj M., Okada Y., Mishra A., Rutten-Jacobs L., Giese A.K., van der Laan S.W., Gretarsdottir S., et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat. Genet. 2018;50:524–537. doi: 10.1038/s41588-018-0058-3. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

