Human lncRNAs harbor conserved modules embedded in different sequence contexts
- PMID: 39040814
- PMCID: PMC11261117
- DOI: 10.1016/j.ncrna.2024.06.013
Human lncRNAs harbor conserved modules embedded in different sequence contexts
Abstract
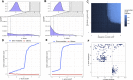
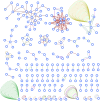
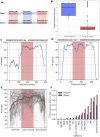
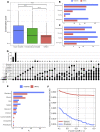
We analyzed the structure of human long non-coding RNA (lncRNAs) genes to investigate whether the non-coding transcriptome is organized in modular domains, as is the case for protein-coding genes. To this aim, we compared all known human lncRNA exons and identified 340 pairs of exons with high sequence and/or secondary structure similarity but embedded in a dissimilar sequence context. We grouped these pairs in 106 clusters based on their reciprocal similarities. These shared modules are highly conserved between humans and the four great ape species, display evidence of purifying selection and likely arose as a result of recent segmental duplications. Our analysis contributes to the understanding of the mechanisms driving the evolution of the non-coding genome and suggests additional strategies towards deciphering the functional complexity of this class of molecules.
Keywords: Non-coding RNAs; Sequence alignments; Structure alignments; lncRNA; lncRNA evolution; lncRNA modules.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures






References
-
- Frankish A., Diekhans M., Jungreis I., Lagarde J., Loveland J.E., Mudge J.M., Sisu C., Wright J.C., Armstrong J., Barnes I., Berry A., Bignell A., Boix C., Carbonell Sala S., Cunningham F., Di Domenico T., Donaldson S., Fiddes I.T., García Girón C., Gonzalez J.M., Grego T., Hardy M., Hourlier T., Howe K.L., Hunt T., Izuogu O.G., Johnson R., Martin F.J., Martínez L., Mohanan S., Muir P., Navarro F.C.P., Parker A., Pei B., Pozo F., Riera F.C., Ruffier M., Schmitt B.M., Stapleton E., Suner M.-M., Sycheva I., Uszczynska-Ratajczak B., Wolf M.Y., Xu J., Yang Y.T., Yates A., Zerbino D., Zhang Y., Choudhary J.S., Gerstein M., Guigó R., Hubbard T.J.P., Kellis M., Paten B., Tress M.L., Flicek P. Gencode 2021. Nucleic Acids Res. 2021;49:D916–D923. - PMC - PubMed
-
- Gao Y., Shang S., Guo S., Li X., Zhou H., Liu H., Sun Y., Wang J., Wang P., Zhi H., Li X., Ning S., Zhang Y. Lnc2Cancer 3.0: an updated resource for experimentally supported lncRNA/circRNA cancer associations and web tools based on RNA-seq and scRNA-seq data. Nucleic Acids Res. 2021;49:D1251–D1258. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

