The phosphatidylethanolamine-binding proteins OsMFT1 and OsMFT2 regulate seed dormancy in rice
- PMID: 39041489
- PMCID: PMC11371141
- DOI: 10.1093/plcell/koae211
The phosphatidylethanolamine-binding proteins OsMFT1 and OsMFT2 regulate seed dormancy in rice
Abstract
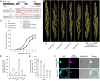
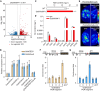
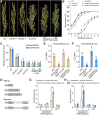
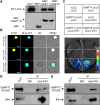
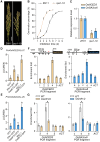
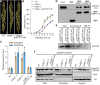
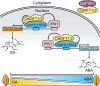
Seed dormancy is crucial for optimal plant life-cycle timing. However, domestication has largely diminished seed dormancy in modern cereal cultivars, leading to challenges such as preharvest sprouting (PHS) and subsequent declines in yield and quality. Therefore, it is imperative to unravel the molecular mechanisms governing seed dormancy for the development of PHS-resistant varieties. In this study, we screened a mutant of BASIC HELIX-LOOP-HELIX TRANSCRIPTION FACTOR4 (OsbHLH004) with decreased seed dormancy and revealed that OsbHLH004 directly regulates the expression of 9-CIS-EPOXYCAROTENOID DIOXYGENASE3 (OsNCED3) and GIBBERELLIN 2-OXIDASE6 (OsGA2ox6) in rice (Oryza sativa). Additionally, we determined that two phosphatidylethanolamine-binding proteins, MOTHER OF FT AND TFL1 and 2 (OsMFT1 and OsMFT2; hereafter OsMFT1/2) interact with OsbHLH004 and Ideal Plant Architecture 1 (IPA1) to regulate their binding capacities on OsNCED3 and OsGA2ox6, thereby promoting seed dormancy. Intriguingly, FT-INTERACTING PROTEIN1 (OsFTIP1) interacts with OsMFT1/2 and affects their nucleocytoplasmic translocation into the nucleus, where OsMFT1/2-OsbHLH004 and OsMFT1/2-IPA1 antagonistically modulate the expression of OsNCED3 and OsGA2ox6. Our findings reveal that OsFTIP1-mediated inhibition of nuclear translocation of OsMFT1/2 and the dynamic transcriptional modulation of OsNCED3 and OsGA2ox6 by OsMFT1/2-OsbHLH004 and OsMFT1/2-IPA1 complexes in seed dormancy in rice.
© The Author(s) 2024. Published by Oxford University Press on behalf of American Society of Plant Biologists. All rights reserved. For commercial re-use, please contact reprints@oup.com for reprints and translation rights for reprints. All other permissions can be obtained through our RightsLink service via the Permissions link on the article page on our site—for further information please contact journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures

References
MeSH terms
Substances
Grants and funding
- 32370379/National Natural Science Foundation of China
- Y2022QC19/Youth Innovation of Chinese Academy of Agricultural Sciences
- LR21C130001/Zhejiang Provincial Natural Science Foundation
- 2021C02063-1/Key Research and Development Program of Zhejiang
- NRF-NRFI2016-02/Singapore National Research Foundation Investigatorship Programme
LinkOut - more resources
Full Text Sources
Research Materials

