Maximum likelihood estimators for colony-forming units
- PMID: 39041814
- PMCID: PMC11371269
- DOI: 10.1128/spectrum.03946-23
Maximum likelihood estimators for colony-forming units
Abstract
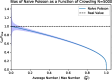
Measuring the abundance of microbes in a sample is a common procedure with a long history, but best practices are not well-conserved across microbiological fields. Serial dilution methods are commonly used to dilute bacterial cultures to produce countable numbers of colonies, and from these counts, to infer bacterial concentrations measured in colony-forming units (CFUs). The most common methods to generate data for CFU point estimates involve plating bacteria on (or in) a solid growth medium and counting their resulting colonies or counting the number of tubes at a given dilution that have growth. Traditionally, these types of data have been analyzed separately using different analytic methods. Here, we build a direct correspondence between these approaches, which allows one to extend the use of the most probable number method from the liquid tubes experiments, for which it was developed, to the growth plates by viewing colony-sized patches of a plate as equivalent to individual tubes. We also discuss how to combine measurements taken at different dilutions, and we review several ways of analyzing colony counts, including the Poisson and truncated Poisson methods. We test all point estimate methods computationally using simulated data. For all methods, we discuss their relevant error bounds, assumptions, strengths, and weaknesses. We provide an online calculator for these estimators.Estimation of the number of microbes in a sample is an important problem with a long history. Yet common practices, such as combining results from different measurements, remain sub-optimal. We provide a comparison of methods for estimating abundance of microbes and detail a mapping between different methods, which allows to extend their range of applicability. This mapping enables higher precision estimates of colony-forming units (CFUs) using the same data already collected for traditional CFU estimation methods. Furthermore, we provide recommendations for how to combine measurements of colony counts taken across dilutions, correcting several misconceptions in the literature.
Keywords: CFU; MPN; bacterial counts; colony count estimation; dilution experiments; dilution plating; maximum likelihood estimator; most probable number.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- McCrady MH. 1915. The numerical interpretation of fermentation-tube results. J Infect Dis 17:183–212. doi: 10.1093/infdis/17.1.183 - DOI
-
- Cochran WG. 1950. Estimation of bacterial densities by means of the "most probable number" Biometrics 6:105–116. - PubMed
-
- Fisher R, Thornton H, Mackenzie W. 1922. The accuracy of the plating method of estimating the density of bacterial populations: with particular reference to the use of thornton’s agar medium with soil samples. Ann Appl Biol 9:325. doi: 10.1111/j.1744-7348.1922.tb05962.x - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

