Resolving the 22q11.2 deletion using CTLR-Seq reveals chromosomal rearrangement mechanisms and individual variance in breakpoints
- PMID: 39042694
- PMCID: PMC11295037
- DOI: 10.1073/pnas.2322834121
Resolving the 22q11.2 deletion using CTLR-Seq reveals chromosomal rearrangement mechanisms and individual variance in breakpoints
Abstract
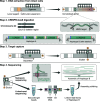
We developed a generally applicable method, CRISPR/Cas9-targeted long-read sequencing (CTLR-Seq), to resolve, haplotype-specifically, the large and complex regions in the human genome that had been previously impenetrable to sequencing analysis, such as large segmental duplications (SegDups) and their associated genome rearrangements. CTLR-Seq combines in vitro Cas9-mediated cutting of the genome and pulse-field gel electrophoresis to isolate intact large (i.e., up to 2,000 kb) genomic regions that encompass previously unresolvable genomic sequences. These targets are then sequenced (amplification-free) at high on-target coverage using long-read sequencing, allowing for their complete sequence assembly. We applied CTLR-Seq to the SegDup-mediated rearrangements that constitute the boundaries of, and give rise to, the 22q11.2 Deletion Syndrome (22q11DS), the most common human microdeletion disorder. We then performed de novo assembly to resolve, at base-pair resolution, the full sequence rearrangements and exact chromosomal breakpoints of 22q11.2DS (including all common subtypes). Across multiple patients, we found a high degree of variability for both the rearranged SegDup sequences and the exact chromosomal breakpoint locations, which coincide with various transposons within the 22q11.2 SegDups, suggesting that 22q11DS can be driven by transposon-mediated genome recombination. Guided by CTLR-Seq results from two 22q11DS patients, we performed three-dimensional chromosomal folding analysis for the 22q11.2 SegDups from patient-derived neurons and astrocytes and found chromosome interactions anchored within the SegDups to be both cell type-specific and patient-specific. Lastly, we demonstrated that CTLR-Seq enables cell-type specific analysis of DNA methylation patterns within the deletion haplotype of 22q11DS.
Keywords: chromosome interactions; copy number variation; genome repeats; microdeletion syndromes; targeted genome assembly.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures





References
MeSH terms
Grants and funding
- R01 MH061675/MH/NIMH NIH HHS/United States
- RSG-13-297-01-TBG/American Cancer Society (ACS)
- R01 HG010359/HG/NHGRI NIH HHS/United States
- P01 HG000205/HG/NHGRI NIH HHS/United States
- P01HG00205ESH/HHS | NIH | National Human Genome Research Institute (NHGRI)
- R01 MH067257/MH/NIMH NIH HHS/United States
- K01MH129758/HHS | NIH | National Institute of Mental Health (NIMH)
- R01 MH059587/MH/NIMH NIH HHS/United States
- U24 MH068457/MH/NIMH NIH HHS/United States
- R01 MH059588/MH/NIMH NIH HHS/United States
- T32 HL110952/HL/NHLBI NIH HHS/United States
- R01 MH060879/MH/NIMH NIH HHS/United States
- R01 HG006137/HG/NHGRI NIH HHS/United States
- Uytengsu-Hamilton 22q11 Neuropsychiatry Research Award/SU | SOM | Stanford Maternal and Child Health Research Institute (MCHRI)
- 1807371/National Science Foundation (NSF)
- R01 MH060870/MH/NIMH NIH HHS/United States
- U01 HG010963/HG/NHGRI NIH HHS/United States
- R01 MH059571/MH/NIMH NIH HHS/United States
- R01 MH059565/MH/NIMH NIH HHS/United States
- P50 HG007735/HG/NHGRI NIH HHS/United States
- U01HG010963/HHS | NIH | National Human Genome Research Institute (NHGRI)
- Instructor K Support Award/SU | SOM | Stanford Maternal and Child Health Research Institute (MCHRI)
- R01 MH059586/MH/NIMH NIH HHS/United States
- R01 MH100900/MH/NIMH NIH HHS/United States
- P50HG00773506/HHS | NIH | National Human Genome Research Institute (NHGRI)
- DP2 MH100010/MH/NIMH NIH HHS/United States
- DP2 MH100010-01/HHS | NIH | National Institute of Mental Health (NIMH)
- R01 MH059566/MH/NIMH NIH HHS/United States
- K01 MH129758/MH/NIMH NIH HHS/United States
- S10 OD023452/OD/NIH HHS/United States
- R01MH100900/HHS | NIH | National Institute of Mental Health (NIMH)
- HG010359/HHS | NIH | National Human Genome Research Institute (NHGRI)
- R01HG006137/HHS | NIH | National Human Genome Research Institute (NHGRI)
LinkOut - more resources
Full Text Sources

