Exploring the conformational landscape of protein kinases
- PMID: 39043011
- PMCID: PMC11694674
- DOI: 10.1016/j.sbi.2024.102890
Exploring the conformational landscape of protein kinases
Abstract
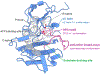
Protein kinases are dynamic enzymes that display complex regulatory mechanisms. Although they possess a structurally conserved catalytic domain, significant conformational dynamics are evident both within a single kinase and across different kinases in the kinome. Here, we highlight methods for exploring this conformational space and its dynamics using kinase domains from ABL1 (Abelson kinase), PKA (protein kinase A), AurA (Aurora A), and PYK2 (proline-rich tyrosine kinase 2) as examples. Such experimental approaches combined with AI-driven methods, such as AlphaFold, will yield discoveries about kinase regulation, the catalytic process, substrate specificity, the effect of disease-associated mutations, as well as new opportunities for structure-based drug design.
Copyright © 2024 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
Figures


References
-
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S: The Protein Kinase Complement of the Human Genome. Science 2002, 298:1912–1934. - PubMed
-
- Lahiry P, Torkamani A, Schork NJ, Hegele RA: Kinase mutations in human disease: interpreting genotype–phenotype relationships. Nat Rev Genet 2010, 11:60–74. - PubMed
-
- Attwood MM, Fabbro D, Sokolov AV, Knapp S, Schiöth HB: Trends in kinase drug discovery: targets, indications and inhibitor design. Nat Rev Drug Discov 2021, 20:839–861. - PubMed
-
- Noble ME, Endicott JA, Johnson LN: Protein Kinase Inhibitors: Insights into Drug Design from Structure. Science 2004, 303:1800–1805. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

