Proposal for a Global Classification and Nomenclature System for A/H9 Influenza Viruses
- PMID: 39043566
- PMCID: PMC11286050
- DOI: 10.3201/eid3008.231176
Proposal for a Global Classification and Nomenclature System for A/H9 Influenza Viruses
Abstract
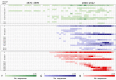
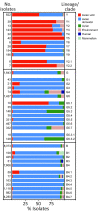
Influenza A/H9 viruses circulate worldwide in wild and domestic avian species, continuing to evolve and posing a zoonotic risk. A substantial increase in human infections with A/H9N2 subtype avian influenza viruses (AIVs) and the emergence of novel reassortants carrying A/H9N2-origin internal genes has occurred in recent years. Different names have been used to describe the circulating and emerging A/H9 lineages. To address this issue, an international group of experts from animal and public health laboratories, endorsed by the WOAH/FAO Network of Expertise on Animal Influenza, has created a practical lineage classification and nomenclature system based on the analysis of 10,638 hemagglutinin sequences from A/H9 AIVs sampled worldwide. This system incorporates phylogenetic relationships and epidemiologic characteristics designed to trace emerging and circulating lineages and clades. To aid in lineage and clade assignment, an online tool has been created. This proposed classification enables rapid comprehension of the global spread and evolution of A/H9 AIVs.
Keywords: A/H9 influenza viruses; Lan Y; Lu L; Pu J; Suggested citation for this article: Fusaro A; Tassoni L; Zhou Y; classification; et al. Proposal for a global classification and nomenclature system for A/H9 influenza viruses. Emerg Infect Dis. 2024 August [date cited]. https://doi.org/10.3201/eid3008.231176; global; hemagglutinin; influenza; nomenclature; phylogeny; respiratory infections; viruses.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

