A multi-classifier system integrated by clinico-histology-genomic analysis for predicting recurrence of papillary renal cell carcinoma
- PMID: 39043664
- PMCID: PMC11266571
- DOI: 10.1038/s41467-024-50369-y
A multi-classifier system integrated by clinico-histology-genomic analysis for predicting recurrence of papillary renal cell carcinoma
Abstract
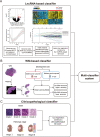
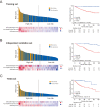
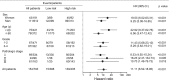
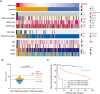
Integrating genomics and histology for cancer prognosis demonstrates promise. Here, we develop a multi-classifier system integrating a lncRNA-based classifier, a deep learning whole-slide-image-based classifier, and a clinicopathological classifier to accurately predict post-surgery localized (stage I-III) papillary renal cell carcinoma (pRCC) recurrence. The multi-classifier system demonstrates significantly higher predictive accuracy for recurrence-free survival (RFS) compared to the three single classifiers alone in the training set and in both validation sets (C-index 0.831-0.858 vs. 0.642-0.777, p < 0.05). The RFS in our multi-classifier-defined high-risk stage I/II and grade 1/2 groups is significantly worse than in the low-risk stage III and grade 3/4 groups (p < 0.05). Our multi-classifier system is a practical and reliable predictor for recurrence of localized pRCC after surgery that can be used with the current staging system to more accurately predict disease course and inform strategies for individualized adjuvant therapy.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

