Identification of potential nucleomodulins of Mycoplasma bovis by direct biotinylation and proximity-based biotinylation approaches
- PMID: 39044956
- PMCID: PMC11263210
- DOI: 10.3389/fmicb.2024.1421585
Identification of potential nucleomodulins of Mycoplasma bovis by direct biotinylation and proximity-based biotinylation approaches
Abstract
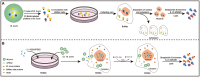
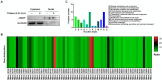
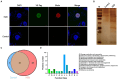
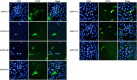
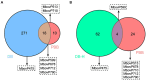
Mycoplasma bovis (M. bovis) is a significant bovine pathogen associated with various diseases, including bovine bronchopneumonia and mastitis resulting in substantial economic losses within the livestock industry. However, the development of effective control measures for M. bovis is hindered by a limited understanding of its virulence factors and pathogenesis. Nucleomodulins are newly identified secreted proteins of bacteria that internalize the host nuclei to regulate host cell gene expression and serve as critical virulence factors. Although recent reports have initiated exploration of mycoplasma nucleomodulins, the efficiency of conventional techniques for identification is very limited. Therefore, this study aimed to establish high-throughput methods to identify novel nucleomodulins of M. bovis. Using a direct biotinylation (DB) approach, a total of 289 proteins were identified including 66 high abundant proteins. In parallel, the use of proximity-based biotinylation (PBB), identified 28 proteins. Finally, seven nucleomodulins were verified to be nuclear by transfecting the bovine macrophage cell line BoMac with the plasmids encoding EGFP-fused proteins and observed with Opera Phenix, including the known nucleomodulin MbovP475 and six novel nucleomodulins. The novel nucleomodulins were four ribosomal proteins (MbovP599, MbovP678, MbovP710, and MbovP712), one transposase (MbovP790), and one conserved hypothetical protein (MbovP513). Among them, one unique nucleomodulin MbovP475 was identified with DB, two unique nucleomodulins (MbovP513 and MbovP710) with PBB, and four nucleomodulins by both. Overall, these findings established a foundation for further research on M. bovis nucleomodulin-host interactions for identification of new virulence factors.
Keywords: Mycoplasma bovis; direct biotinylation; nucleomodulin; proximity-based biotinylation; secreted proteins.
Copyright © 2024 Lu, Chen, Zhang, Fu, Raheem, Chen, Chen, Hu, Chen, Schieck, Zhao and Guo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Miscellaneous

