Viruses of free-roaming and hunting dogs in Uganda show elevated prevalence, richness and abundance across a gradient of contact with wildlife
- PMID: 39045787
- PMCID: PMC11316573
- DOI: 10.1099/jgv.0.002011
Viruses of free-roaming and hunting dogs in Uganda show elevated prevalence, richness and abundance across a gradient of contact with wildlife
Abstract
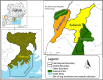
Domestic dogs (Canis lupus familiaris) live with humans, frequently contact other animals and may serve as intermediary hosts for the transmission of viruses. Free-roaming dogs, which account for over 70% of the world's domestic dog population, may pose a particularly high risk in this regard. We conducted an epidemiological study of dog viromes in three locations in Uganda, representing low, medium and high rates of contact with wildlife, ranging from dogs owned specifically for traditional hunting in a biodiversity and disease 'hotspot' to pets in an affluent suburb. We quantified rates of contact between dogs and wildlife through owner interviews and conducted canine veterinary health assessments. We then applied broad-spectrum viral metagenomics to blood plasma samples, from which we identified 46 viruses, 44 of which were previously undescribed, in three viral families, Sedoreoviridae, Parvoviridae and Anelloviridae. All 46 viruses (100 %) occurred in the high-contact population of dogs compared to 63 % and 39 % in the medium- and low-contact populations, respectively. Viral prevalence ranged from 2.1 % to 92.0 % among viruses and was highest, on average, in the high-contact population (22.3 %), followed by the medium-contact (12.3 %) and low-contact (4.8 %) populations. Viral richness (number of viruses per dog) ranged from 0 to 27 and was markedly higher, on average, in the high-contact population (10.2) than in the medium-contact (5.7) or low-contact (2.3) populations. Viral richness was strongly positively correlated with the number of times per year that a dog was fed wildlife and negatively correlated with the body condition score, body temperature and packed cell volume. Viral abundance (cumulative normalized metagenomic read density) varied 124-fold among dogs and was, on average, 4.1-fold higher and 2.4-fold higher in the high-contact population of dogs than in the low-contact or medium-contact populations, respectively. Viral abundance was also strongly positively correlated with the number of times per year that a dog was fed wildlife, negatively correlated with packed cell volume and positively correlated with white blood cell count. These trends were driven by nine viruses in the family Anelloviridae, genus Thetatorquevirus, and by one novel virus in the family Sedoreoviridae, genus Orbivirus. The genus Orbivirus contains zoonotic viruses and viruses that dogs can acquire through ingestion of infected meat. Overall, our findings show that viral prevalence, richness and abundance increased across a gradient of contact between dogs and wildlife and that the health status of the dog modified viral infection. Other ecological, geographic and social factors may also have contributed to these trends. Our finding of a novel orbivirus in dogs with high wildlife contact supports the idea that free-roaming dogs may serve as intermediary hosts for viruses of medical importance to humans and other animals.
Keywords: Africa; Uganda; ecology; epidemiology; free-roaming dogs; viruses; zoonoses.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Falls prevention interventions for community-dwelling older adults: systematic review and meta-analysis of benefits, harms, and patient values and preferences.Syst Rev. 2024 Nov 26;13(1):289. doi: 10.1186/s13643-024-02681-3. Syst Rev. 2024. PMID: 39593159 Free PMC article.
-
Sexual Harassment and Prevention Training.2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36508513 Free Books & Documents.
-
Population-based biomedical sexually transmitted infection control interventions for reducing HIV infection.Cochrane Database Syst Rev. 2011 Mar 16;(3):CD001220. doi: 10.1002/14651858.CD001220.pub3. Cochrane Database Syst Rev. 2011. PMID: 21412869
-
The effect of sample site and collection procedure on identification of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Dec 16;12(12):CD014780. doi: 10.1002/14651858.CD014780. Cochrane Database Syst Rev. 2024. PMID: 39679851 Free PMC article.
-
Biases and complementarity in gut viromes obtained from bulk and virus-like particle-enriched metagenomic sequencing.Microbiol Spectr. 2025 Aug 5;13(8):e0001325. doi: 10.1128/spectrum.00013-25. Epub 2025 Jul 2. Microbiol Spectr. 2025. PMID: 40600714 Free PMC article.
Cited by
-
Prevalence of common conditions and associated mortalities of dogs treated at the small animal clinic, Makerere University, Kampala, Uganda.BMC Vet Res. 2024 Dec 31;20(1):590. doi: 10.1186/s12917-024-04432-x. BMC Vet Res. 2024. PMID: 39736747 Free PMC article.
References
-
- Gompper ME. In: Free-Ranging Dogs and Wildlife Conservation. Gompper ME, editor. New York: Oxford University Press; 2013. The dog–human–wildlife interface: assessing the scope of the problem; pp. 9–54. - DOI
-
- Hughes J, Macdonald DW. A review of the interactions between free-roaming domestic dogs and wildlife. Biol Conserv. 2013;157:341–351. doi: 10.1016/j.biocon.2012.07.005. - DOI
-
- Darvishi M, Omrani nava A, Karimi E, Nouri M, Meigooni SS, et al. Human and animal bites. Caspian J Environ Sci. 2023;21:445–456.
-
- Stauffer K, Wallace R, Galland GG, Marano N. In: CDC Yellow Book. Nemhauser J, editor. Atlanta, Georgia: Centers for Disease Control and Prevention; 2024. Zoonotic exposures: bites, stings, scratches & other hazards; p. 4. p.
-
- Lino S, Rossa M, Fernandes JM, Barros T, Lino A, et al. Dog in sheep’s clothing: livestock depredation by free-ranging dogs may pose new challenges to wolf conservation. Eur J Wildl Res. 2023;69:107. doi: 10.1007/s10344-023-01740-9. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

