Strigolactone-induced degradation of SMXL7 and SMXL8 contributes to gibberellin- and auxin-mediated fiber cell elongation in cotton
- PMID: 39046066
- PMCID: PMC11371155
- DOI: 10.1093/plcell/koae212
Strigolactone-induced degradation of SMXL7 and SMXL8 contributes to gibberellin- and auxin-mediated fiber cell elongation in cotton
Abstract
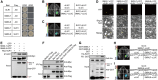
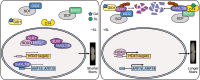
Cotton (Gossypium) fiber length, a key trait determining fiber yield and quality, is highly regulated by a class of recently identified phytohormones, strigolactones (SLs). However, the underlying molecular mechanisms of SL signaling involved in fiber cell development are largely unknown. Here, we show that the SL signaling repressors MORE AXILLARY GROWTH2-LIKE7 (GhSMXL7) and GhSMXL8 negatively regulate cotton fiber elongation. Specifically, GhSMXL7 and GhSMXL8 inhibit the polyubiquitination and degradation of the gibberellin (GA)-triggered DELLA protein (GhSLR1). Biochemical analysis revealed that GhSMXL7 and GhSMXL8 physically interact with GhSLR1, which interferes with the association of GhSLR1 with the E3 ligase GA INSENSITIVE2 (GhGID2), leading to the repression of GA signal transduction. GhSMXL7 also interacts with the transcription factor GhHOX3, preventing its binding to the promoters of essential fiber elongation regulatory genes. Moreover, both GhSMXL7 and GhSMXL8 directly bind to the promoter regions of the AUXIN RESPONSE FACTOR (ARF) genes GhARF18-10A, GhARF18-10D, and GhARF19-7D to suppress their expression. Cotton plants in which GhARF18-10A, GhARF18-10D, and GhARF19-7D transcript levels had been reduced by virus-induced gene silencing (VIGS) displayed reduced fiber length compared with control plants. Collectively, our findings reveal a mechanism illustrating how SL integrates GA and auxin signaling to coordinately regulate plant cell elongation at the single-cell level.
© The Author(s) 2024. Published by Oxford University Press on behalf of American Society of Plant Biologists. All rights reserved. For commercial re-use, please contact reprints@oup.com for reprints and translation rights for reprints. All other permissions can be obtained through our RightsLink service via the Permissions link on the article page on our site—for further information please contact journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest statement. The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

