KDM6A regulates immune response genes in multiple myeloma
- PMID: 39046770
- PMCID: PMC11952010
- DOI: 10.1182/blood.2024024518
KDM6A regulates immune response genes in multiple myeloma
Abstract
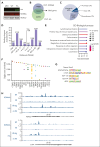
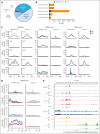
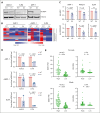
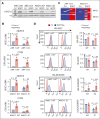
The histone H3 at lysine 27 (H3K27) demethylase lysine demethylase 6A (KDM6A) is a tumor suppressor in multiple cancers, including multiple myeloma (MM). We created isogenic MM cells disrupted for KDM6A and tagged the endogenous protein to facilitate genome-wide studies. KDM6A binds genes associated with immune recognition and cytokine signaling. Most importantly, KDM6A binds and activates NLRC5 and CIITA, which encode regulators of major histocompatibility complex genes. Patient data indicate that NLRC5 and CIITA are downregulated in MM with low KDM6A expression. Chromatin analysis shows that KDM6A binds poised and active enhancers and KDM6A loss led to decreased H3K27ac at enhancers, increased H3K27me3 levels in body of genes bound by KDM6A, and decreased gene expression. Reestablishing histone acetylation with an HDAC3 inhibitor leads to upregulation of major histocompatibility complex expression, offering a strategy to restore immunogenicity of KDM6A-deficient tumors. Loss of Kdm6a in Kirsten rat sarcoma virus (K-RAS)-transformed murine fibroblasts led to increased growth in vivo associated with decreased T-cell infiltration.
© 2024 American Society of Hematology. Published by Elsevier Inc. All rights are reserved, including those for text and data mining, AI training, and similar technologies.
Conflict of interest statement
Conflict-of-interest disclosure: J.D.L. reports research support from Epizyme/Ipsen; and a consultancy role at AstraZeneca. L.H.B. reports a consultancy role at AstraZeneca; C.S.M. serves on the scientific advisory board of Adicet Bio; reports consultant/honoraria from Genentech, Nerviano, Secura Bio, and Oncopeptides; and research funding from EMD Serono, Karyopharm, Sanofi, Nurix, Bristol Myers Squibb, H3 Biomedicine/Eisai, Springworks, Abcuro, Novartis, and Opna. The remaning authors declare no competing financial interests.
Figures


Update of
-
KDM6A Regulates Immune Response Genes in Multiple Myeloma.bioRxiv [Preprint]. 2024 Feb 12:2024.02.12.579179. doi: 10.1101/2024.02.12.579179. bioRxiv. 2024. Update in: Blood. 2024 Oct 3;144(14):1508-1520. doi: 10.1182/blood.2024024518. PMID: 38405853 Free PMC article. Updated. Preprint.
Comment in
-
KDM6A controls immunogenicity in MM.Blood. 2024 Oct 3;144(14):1465-1467. doi: 10.1182/blood.2024026013. Blood. 2024. PMID: 39361299 No abstract available.
References
-
- Calasanz MJ, Cigudosa JC, Odero MD, et al. Cytogenetic analysis of 280 patients with multiple myeloma and related disorders: primary breakpoints and clinical correlations. Genes Chromosomes Cancer. 1997;18(2):84–93. - PubMed
-
- Dewald GW, Kyle RA, Hicks GA, Greipp PR. The clinical significance of cytogenetic studies in 100 patients with multiple myeloma, plasma cell leukemia, or amyloidosis. Blood. 1985;66(2):380–390. - PubMed
-
- Sawyer JR, Waldron JA, Jagannath S, Barlogie B. Cytogenetic findings in 200 patients with multiple myeloma. Cancer Genet Cytogenet. 1995;82(1):41–49. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

