Measurement of solubility product reveals the interplay of oligomerization and self-association for defining condensate formation
- PMID: 39046778
- PMCID: PMC11449392
- DOI: 10.1091/mbc.E24-01-0030
Measurement of solubility product reveals the interplay of oligomerization and self-association for defining condensate formation
Abstract
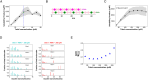
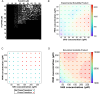
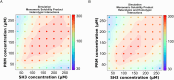
Cellular condensates often consist of 10s to 100s of distinct interacting molecular species. Because of the complexity of these interactions, predicting the point at which they will undergo phase separation is daunting. Using experiments and computation, we therefore studied a simple model system consisting of polySH3 and polyPRM designed for pentavalent heterotypic binding. We tested whether the peak solubility product, or the product of the dilute phase concentration of each component, is a predictive parameter for the onset of phase separation. Titrating up equal total concentrations of each component showed that the maximum solubility product does approximately coincide with the threshold for phase separation in both experiments and models. However, we found that measurements of dilute phase concentration include small oligomers and monomers; therefore, a quantitative comparison of the experiments and models required inclusion of small oligomers in the model analysis. Even with the inclusion of small polyPRM and polySH3 oligomers, models did not predict experimental results. This led us to perform dynamic light scattering experiments, which revealed homotypic binding of polyPRM. Addition of this interaction to our model recapitulated the experimentally observed asymmetry. Thus, comparing experiments with simulation reveals that the solubility product can be predictive of the interactions underlying phase separation, even if small oligomers and low affinity homotypic interactions complicate the analysis.
Conflict of interest statement
Conflict of interest: The authors declare a financial conflict of interest.
Figures



Update of
-
Measurement of solubility product in a model condensate reveals the interplay of small oligomerization and self-association.bioRxiv [Preprint]. 2024 Jan 26:2024.01.23.576869. doi: 10.1101/2024.01.23.576869. bioRxiv. 2024. Update in: Mol Biol Cell. 2024 Sep 1;35(9):ar122. doi: 10.1091/mbc.E24-01-0030. PMID: 38328089 Free PMC article. Updated. Preprint.
References
-
- Alberti S, Hyman AA (2021). Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat Rev Mol Cell Biol 22, 196–213. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

