Exploring the Macroevolutionary Signature of Asymmetric Inheritance at Speciation
- PMID: 39046782
- PMCID: PMC11958934
- DOI: 10.1093/sysbio/syae043
Exploring the Macroevolutionary Signature of Asymmetric Inheritance at Speciation
Abstract
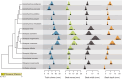
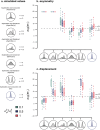
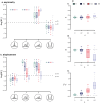
Popular comparative phylogenetic models such as Brownian Motion, Ornstein-Ulhenbeck, and their extensions assume that, at speciation, a trait value is inherited identically by 2 descendant species. This assumption contrasts with models of speciation at a micro-evolutionary scale where descendants' phenotypic distributions are sub-samples of the ancestral distribution. Different speciation mechanisms can lead to a displacement of the ancestral phenotypic mean among descendants and an asymmetric inheritance of the ancestral phenotypic variance. In contrast, even macro-evolutionary models that account for intraspecific variance assume symmetrically conserved inheritance of ancestral phenotypic distribution at speciation. Here, we develop an Asymmetric Brownian Motion model (ABM) that relaxes the assumption of symmetric and conserved inheritance of the ancestral distribution at the time of speciation. The ABM jointly models the evolution of both intra- and inter-specific phenotypic variation. It also infers the mode of phenotypic inheritance at speciation, which can range from a symmetric and conserved inheritance, where descendants inherit the ancestral distribution, to an asymmetric and displaced inheritance, where descendants inherit divergent phenotypic means and variances. To demonstrate this model, we analyze the evolution of beak morphology in Darwin finches, finding evidence of displacement at speciation. The ABM model helps to bridge micro- and macro-evolutionary models of trait evolution by providing a more robust framework for testing the effects of ecological speciation, character displacement, and niche partitioning on trait evolution at the macro-evolutionary scale.
Keywords: Character displacement; phenotypic evolution; phylogenetic comparative methods; speciation.
© The Author(s) 2024. Published by Oxford University Press, on behalf of the Society of Systematic Biologists.
Conflict of interest statement
None declared.
Figures


References
-
- Abzhanov A. 2017. The old and new faces of morphology: the legacy of D’Arcy Thompson’s “theory of transformations” and “laws of growth”. Development (Cambridge) 144:4284–4297. - PubMed
-
- Ackermann M., Doebeli M.. 2004. Evolution of niche width and adaptive diversification. Evolution 58:2599–2612. - PubMed
-
- Beaulieu J.M., Jhwueng D.-C., Boettiger C., O’Meara B.C.. 2012. Modeling stabilizing selection: expanding the Ornstein-Uhlenbeck model of adaptive evolution. Evolution 66:2369–2383. - PubMed
-
- Blomberg S.P., Garland T., Ives A.R.. 2003. Signal in comparative data: testing for phylogenetic behavioral traits are more labile. Evolution 57:717–745. - PubMed
-
- Bokma F. 2002. Detection of punctuated equilibrium from molecular phylogenies. J. Evol. Biol. 15:1048–1056.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

