Control of Solid-Supported Intra- vs Interstrand Stille Coupling Reactions for Synthesis of DNA-Oligophenylene Conjugates
- PMID: 39046902
- PMCID: PMC11342295
- DOI: 10.1021/acs.bioconjchem.4c00310
Control of Solid-Supported Intra- vs Interstrand Stille Coupling Reactions for Synthesis of DNA-Oligophenylene Conjugates
Abstract
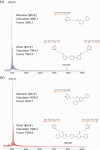
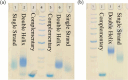
Programmed DNA structures and assemblies are readily accessible, but site-specific functionalization is critical to realize applications in various fields such as nanoelectronics, nanomaterials and biomedicine. Besides pre- and post-DNA synthesis conjugation strategies, on-solid support reactions offer advantages in certain circumstances. We describe on-solid support internucleotide coupling reactions, often considered undesirable, and a workaround strategy to overcome them. Palladium coupling reactions enabled on-solid support intra- and interstrand coupling between single-stranded DNAs (ss-DNAs). Dilution with a capping agent suppressed interstrand coupling, maximizing intrastrand coupling. Alternatively, interstrand coupling actually proved advantageous to provide dimeric organic/DNA conjugates that could be conveniently separated from higher oligomers, and was more favorable with longer terphenyl coupling partners.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
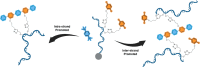



References
-
- Seeman N. C.Structural DNA Nanotechnology; Cambridge University Press: Cambridge, UK, 2015.
-
- Seeman N. C.; Sleiman H. F. DNA nanotechnology. Nature Reviews Materials 2018, 3, 17068.10.1038/natrevmats.2017.68. - DOI
-
- Herdewijn P.Oligonucleotide Synthesis: Methods and Applications; Humana Press: Totowa, NJ, 2005; p xvi, 435 p.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

