FedGMMAT: Federated generalized linear mixed model association tests
- PMID: 39047024
- PMCID: PMC11299833
- DOI: 10.1371/journal.pcbi.1012142
FedGMMAT: Federated generalized linear mixed model association tests
Abstract
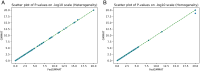
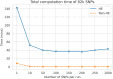
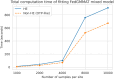
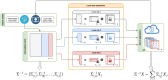
Increasing genetic and phenotypic data size is critical for understanding the genetic determinants of diseases. Evidently, establishing practical means for collaboration and data sharing among institutions is a fundamental methodological barrier for performing high-powered studies. As the sample sizes become more heterogeneous, complex statistical approaches, such as generalized linear mixed effects models, must be used to correct for the confounders that may bias results. On another front, due to the privacy concerns around Protected Health Information (PHI), genetic information is restrictively protected by sharing according to regulations such as Health Insurance Portability and Accountability Act (HIPAA). This limits data sharing among institutions and hampers efforts around executing high-powered collaborative studies. Federated approaches are promising to alleviate the issues around privacy and performance, since sensitive data never leaves the local sites. Motivated by these, we developed FedGMMAT, a federated genetic association testing tool that utilizes a federated statistical testing approach for efficient association tests that can correct for confounding fixed and additive polygenic random effects among different collaborating sites. Genetic data is never shared among collaborating sites, and the intermediate statistics are protected by encryption. Using simulated and real datasets, we demonstrate FedGMMAT can achieve the virtually same results as pooled analysis under a privacy-preserving framework with practical resource requirements.
Copyright: © 2024 Li et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Secure and federated genome-wide association studies for biobank-scale datasets.Nat Genet. 2025 Apr;57(4):809-814. doi: 10.1038/s41588-025-02109-1. Epub 2025 Feb 24. Nat Genet. 2025. PMID: 39994472 Free PMC article.
-
Doctors Routinely Share Health Data Electronically Under HIPAA, and Sharing With Patients and Patients' Third-Party Health Apps is Consistent: Interoperability and Privacy Analysis.J Med Internet Res. 2020 Sep 2;22(9):e19818. doi: 10.2196/19818. J Med Internet Res. 2020. PMID: 32876582 Free PMC article.
-
A novel privacy-preserving federated genome-wide association study framework and its application in identifying potential risk variants in ankylosing spondylitis.Brief Bioinform. 2021 May 20;22(3):bbaa090. doi: 10.1093/bib/bbaa090. Brief Bioinform. 2021. PMID: 32591779
-
Federated Analysis of Neuroimaging Data: A Review of the Field.Neuroinformatics. 2022 Apr;20(2):377-390. doi: 10.1007/s12021-021-09550-7. Epub 2021 Nov 22. Neuroinformatics. 2022. PMID: 34807353 Free PMC article. Review.
-
Sharing sensitive data in life sciences: an overview of centralized and federated approaches.Brief Bioinform. 2024 May 23;25(4):bbae262. doi: 10.1093/bib/bbae262. Brief Bioinform. 2024. PMID: 38836701 Free PMC article. Review.
Cited by
-
Modeling the Dependence Structure in Genome Wide Association Studies of Binary Phenotypes in Family Data.Behav Genet. 2020 Nov;50(6):423-439. doi: 10.1007/s10519-020-10010-2. Epub 2020 Aug 17. Behav Genet. 2020. PMID: 32804302 Free PMC article.
-
Clarifying the Risk of Lung Disease in SZ Alpha-1 Antitrypsin Deficiency.Am J Respir Crit Care Med. 2020 Jul 1;202(1):73-82. doi: 10.1164/rccm.202002-0262OC. Am J Respir Crit Care Med. 2020. PMID: 32197047 Free PMC article.
-
A One-Shot Lossless Algorithm for Cross-Cohort Learning in Mixed-Outcomes Analysis.medRxiv [Preprint]. 2025 Apr 29:2024.01.09.24301073. doi: 10.1101/2024.01.09.24301073. medRxiv. 2025. PMID: 38260403 Free PMC article. Preprint.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

