Iron regulatory protein 2 contributes to antimicrobial immunity by preserving lysosomal function in macrophages
- PMID: 39047035
- PMCID: PMC11295080
- DOI: 10.1073/pnas.2321929121
Iron regulatory protein 2 contributes to antimicrobial immunity by preserving lysosomal function in macrophages
Abstract
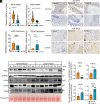
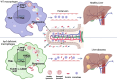
Colorectal cancer and Crohn's disease patients develop pyogenic liver abscesses due to failures of immune cells to fight off bacterial infections. Here, we show that mice lacking iron regulatory protein 2 (Irp2), globally (Irp2-/-) or myeloid cell lineage (Lysozyme 2 promoter-driven, LysM)-specifically (Irp2ΔLysM), are highly susceptible to liver abscesses when the intestinal tissue was injured with dextran sodium sulfate treatment. Further studies demonstrated that Irp2 is required for lysosomal acidification and biogenesis, both of which are crucial for bacterial clearance. In Irp2-deficient liver tissue or macrophages, the nuclear location of transcription factor EB (Tfeb) was remarkably reduced, leading to the downregulation of Tfeb target genes that encode critical components for lysosomal biogenesis. Tfeb mislocalization was reversed by hypoxia-inducible factor 2 inhibitor PT2385 and, independently, through inhibition of lactic acid production. These experimental findings were confirmed clinically in patients with Crohn's disease and through bioinformatic searches in databases from Crohn's disease or ulcerative colitis biopsies showing loss of IRP2 and transcription factor EB (TFEB)-dependent lysosomal gene expression. Overall, our study highlights a mechanism whereby Irp2 supports nuclear translocation of Tfeb and lysosomal function, preserving macrophage antimicrobial activity and protecting the liver against invading bacteria during intestinal inflammation.
Keywords: HIF2; IRP2; TFEB localization; lactic acid; lysosomal function.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures






References
-
- Lin J. N., et al. , Pyogenic liver abscess in patients with inflammatory bowel disease: A nationwide cohort study. Liver Int. 36, 136–144 (2016). - PubMed
MeSH terms
Substances
Grants and funding
- 32271221/MOST | National Natural Science Foundation of China (NSFC)
- 82170592/MOST | National Natural Science Foundation of China (NSFC)
- 2023TQ0154/China Postdoctoral Science Foundation (China Postdoctoral Foundation Project)
- 2023M741647/China Postdoctoral Science Foundation (China Postdoctoral Foundation Project)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

