Analyzing embryo dormancy at single-cell resolution reveals dynamic transcriptional responses and activation of integrin-Yap/Taz prosurvival signaling
- PMID: 39047740
- PMCID: PMC7617458
- DOI: 10.1016/j.stem.2024.06.015
Analyzing embryo dormancy at single-cell resolution reveals dynamic transcriptional responses and activation of integrin-Yap/Taz prosurvival signaling
Abstract
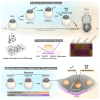
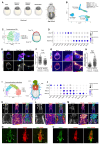
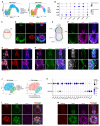
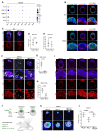
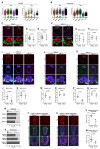
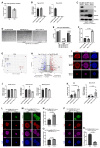
Embryonic diapause is a reproductive adaptation that enables some mammalian species to halt the otherwise continuous pace of embryonic development. In this dormant state, the embryo exploits poorly understood regulatory mechanisms to preserve its developmental potential for prolonged periods of time. Here, using mouse embryos and single-cell RNA sequencing, we molecularly defined embryonic diapause at single-cell resolution, revealing transcriptional dynamics while the embryo seemingly resides in a state of suspended animation. Additionally, we found that the dormant pluripotent cells rely on integrin receptors to sense their microenvironment and preserve their viability via Yap/Taz-mediated prosurvival signaling.
Keywords: Taz; Yap; embryo dormancy; embryonic diapause; embryonic stem cells; epiblast; extracellular matrix; integrin; pluripotency; single-cell RNA sequencing.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

Similar articles
-
Evaluating the involvement and mutual interaction of wbp2 and yap in embryogenesis with an emphasis on liver function in zebrafish embryos.Tissue Cell. 2024 Dec;91:102600. doi: 10.1016/j.tice.2024.102600. Epub 2024 Oct 29. Tissue Cell. 2024. PMID: 39486132
-
Wnt/β-Catenin Signaling Regulates Yap/Taz Activity during Embryonic Development in Zebrafish.Int J Mol Sci. 2024 Sep 17;25(18):10005. doi: 10.3390/ijms251810005. Int J Mol Sci. 2024. PMID: 39337493 Free PMC article.
-
Integrin signalling regulates YAP and TAZ to control skin homeostasis.Development. 2016 May 15;143(10):1674-87. doi: 10.1242/dev.133728. Epub 2016 Mar 17. Development. 2016. PMID: 26989177 Free PMC article.
-
YAP/TAZ Signalling Controls Epidermal Keratinocyte Fate.Int J Mol Sci. 2024 Nov 30;25(23):12903. doi: 10.3390/ijms252312903. Int J Mol Sci. 2024. PMID: 39684613 Free PMC article. Review.
-
Role of the Hippo-YAP/TAZ Pathway in Epithelioid Hemangioendothelioma and its Potential as a Therapeutic Target.Anticancer Res. 2024 Oct;44(10):4147-4153. doi: 10.21873/anticanres.17245. Anticancer Res. 2024. PMID: 39348982 Review.
Cited by
-
Protocol for induction, maintenance, and exit from embryo dormancy in mice.STAR Protoc. 2025 Jun 20;6(2):103813. doi: 10.1016/j.xpro.2025.103813. Epub 2025 May 6. STAR Protoc. 2025. PMID: 40333191 Free PMC article.
References
-
- Renfree MB, Shaw G. Diapause. Annu Rev Physiol. 2000;62:353–375. - PubMed
-
- Yoshinaga K, Adams CE. Delayed implantation in the spayed, progesterone treated adult mouse. J Reprod Fertil. 1966;12:593–595. - PubMed
-
- Fan R, Kim YS, Wu J, Chen R, Zeuschner D, Mildner K, Adachi K, Wu G, Galatidou S, Li J, et al. Wnt/beta-catenin/Esrrb signalling controls the tissue-scale reorganization and maintenance of the pluripotent lineage during murine embryonic diapause. Nat Commun. 2020;11:5499. doi: 10.1038/s41467-020-19353-0. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

