A ligand discovery toolbox for the WWE domain family of human E3 ligases
- PMID: 39048679
- PMCID: PMC11269756
- DOI: 10.1038/s42003-024-06584-w
A ligand discovery toolbox for the WWE domain family of human E3 ligases
Abstract
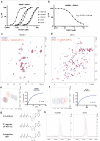
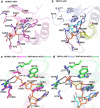
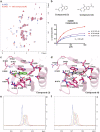
The WWE domain is a relatively under-researched domain found in twelve human proteins and characterized by a conserved tryptophan-tryptophan-glutamate (WWE) sequence motif. Six of these WWE domain-containing proteins also contain domains with E3 ubiquitin ligase activity. The general recognition of poly-ADP-ribosylated substrates by WWE domains suggests a potential avenue for development of Proteolysis-Targeting Chimeras (PROTACs). Here, we present novel crystal structures of the HUWE1, TRIP12, and DTX1 WWE domains in complex with PAR building blocks and their analogs, thus enabling a comprehensive analysis of the PAR binding site structural diversity. Furthermore, we introduce a versatile toolbox of biophysical and biochemical assays for the discovery and characterization of novel WWE domain binders, including fluorescence polarization-based PAR binding and displacement assays, 15N-NMR-based binding affinity assays and 19F-NMR-based competition assays. Through these assays, we have characterized the binding of monomeric iso-ADP-ribose (iso-ADPr) and its nucleotide analogs with the aforementioned WWE proteins. Finally, we have utilized the assay toolbox to screen a small molecule fragment library leading to the successful discovery of novel ligands targeting the HUWE1 WWE domain.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

