A human autoimmune organoid model reveals IL-7 function in coeliac disease
- PMID: 39048815
- PMCID: PMC11747932
- DOI: 10.1038/s41586-024-07716-2
A human autoimmune organoid model reveals IL-7 function in coeliac disease
Abstract
In vitro models of autoimmunity are constrained by an inability to culture affected epithelium alongside the complex tissue-resident immune microenvironment. Coeliac disease (CeD) is an autoimmune disease in which dietary gluten-derived peptides bind to the major histocompatibility complex (MHC) class II human leukocyte antigen molecules (HLA)-DQ2 or HLA-DQ8 to initiate immune-mediated duodenal mucosal injury1-4. Here, we generated air-liquid interface (ALI) duodenal organoids from intact fragments of endoscopic biopsies that preserve epithelium alongside native mesenchyme and tissue-resident immune cells as a unit without requiring reconstitution. The immune diversity of ALI organoids spanned T cells, B and plasma cells, natural killer (NK) cells and myeloid cells, with extensive T-cell and B-cell receptor repertoires. HLA-DQ2.5-restricted gluten peptides selectively instigated epithelial destruction in HLA-DQ2.5-expressing organoids derived from CeD patients, and this was antagonized by blocking MHC-II or NKG2C/D. Gluten epitopes stimulated a CeD organoid immune network response in lymphoid and myeloid subsets alongside anti-transglutaminase 2 (TG2) autoantibody production. Functional studies in CeD organoids revealed that interleukin-7 (IL-7) is a gluten-inducible pathogenic modulator that regulates CD8+ T-cell NKG2C/D expression and is necessary and sufficient for epithelial destruction. Furthermore, endogenous IL-7 was markedly upregulated in patient biopsies from active CeD compared with remission disease from gluten-free diets, predominantly in lamina propria mesenchyme. By preserving the epithelium alongside diverse immune populations, this human in vitro CeD model recapitulates gluten-dependent pathology, enables mechanistic investigation and establishes a proof of principle for the organoid modelling of autoimmunity.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
C.J.K. and A.J.M.S. are inventors on patent WO 2020/247528 describing methods and uses of patient-derived celiac intestinal organoids. C.J.K. and M.M.D. are founders of Mozart Therapeutics and NextVivo, Inc. L.M.S has been a consultant during the last 3 years for BMS, GSK, Mozart Therapeutics, Ono Pharmaceutical, Precigen ActoBio, Sanofi-Aventis, SQZ Biotech, Takeda and Topas Therapeutics. All other authors declare no competing interests.
Figures



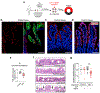





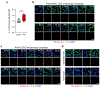

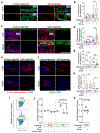



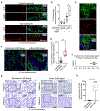
Comment in
-
Organoids identify a role for IL-7 in coeliac disease.Nat Rev Drug Discov. 2024 Oct;23(10):741. doi: 10.1038/d41573-024-00143-y. Nat Rev Drug Discov. 2024. PMID: 39251735 No abstract available.
References
-
- Catassi C, Verdu EF, Bai JC & Lionetti E Coeliac disease. Lancet 399, 2413–2426 (2022). - PubMed
-
- Iversen R & Sollid LM The Immunobiology and Pathogenesis of Celiac Disease. Annu Rev Pathol 18, 47–70 (2023). - PubMed
-
- Marsh MN Gluten, major histocompatibility complex, and the small intestine. A molecular and immunobiologic approach to the spectrum of gluten sensitivity (‘celiac sprue’). Gastroenterology 102, 330–354 (1992). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

