SMYD5 methylation of rpL40 links ribosomal output to gastric cancer
- PMID: 39048817
- PMCID: PMC11625416
- DOI: 10.1038/s41586-024-07718-0
SMYD5 methylation of rpL40 links ribosomal output to gastric cancer
Abstract
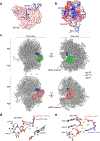
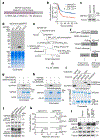
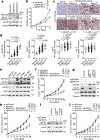
Dysregulated transcription due to disruption in histone lysine methylation dynamics is an established contributor to tumorigenesis1,2. However, whether analogous pathologic epigenetic mechanisms act directly on the ribosome to advance oncogenesis is unclear. Here we find that trimethylation of the core ribosomal protein L40 (rpL40) at lysine 22 (rpL40K22me3) by the lysine methyltransferase SMYD5 regulates mRNA translation output to promote malignant progression of gastric adenocarcinoma (GAC) with lethal peritoneal ascites. A biochemical-proteomics strategy identifies the monoubiquitin fusion protein partner rpL40 (ref. 3) as the principal physiological substrate of SMYD5 across diverse samples. Inhibiting the SMYD5-rpL40K22me3 axis in GAC cell lines reprogrammes protein synthesis to attenuate oncogenic gene expression signatures. SMYD5 and rpL40K22me3 are upregulated in samples from patients with GAC and negatively correlate with clinical outcomes. SMYD5 ablation in vivo in familial and sporadic mouse models of malignant GAC blocks metastatic disease, including peritoneal carcinomatosis. Suppressing SMYD5 methylation of rpL40 inhibits human cancer cell and patient-derived GAC xenograft growth and renders them hypersensitive to inhibitors of PI3K and mTOR. Finally, combining SMYD5 depletion with PI3K-mTOR inhibition and chimeric antigen receptor T cell administration cures an otherwise lethal in vivo mouse model of aggressive GAC-derived peritoneal carcinomatosis. Together, our work uncovers a ribosome-based epigenetic mechanism that facilitates the evolution of malignant GAC and proposes SMYD5 targeting as part of a potential combination therapy to treat this cancer.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Figures












References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

