Developing benzylisoquinoline alkaloid-enriched opium poppy via CRISPR-directed genome editing: A review
- PMID: 39048937
- PMCID: PMC11267691
- DOI: 10.1186/s12870-024-05412-x
Developing benzylisoquinoline alkaloid-enriched opium poppy via CRISPR-directed genome editing: A review
Abstract
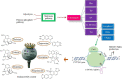
Among plant-derived secondary metabolites are benzylisoquinoline alkaloids (BIAs) that play a vital role in medicine. The most conspicuous BIAs frequently found in opium poppy are morphine, codeine, thebaine, papaverine, sanguinarine, and noscapine. BIAs have provided abundant clinically useful drugs used in the treatment of various diseases and ailments With an increasing demand for these herbal remedies, genetic improvement of poppy plants appears to be essential to live up to the expectations of the pharmaceutical industry. With the advent of clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated9 (Cas9), the field of metabolic engineering has undergone a paradigm shift in its approach due to its appealing attributes, such as the transgene-free editing capability, precision, selectivity, robustness, and versatility. The potentiality of the CRISPR system for manipulating metabolic pathways in opium poppy was demonstrated, but further investigations regarding the use of CRISPR in BIA pathway engineering should be undertaken to develop opium poppy into a bioreactor synthesizing BIAs at the industrial-scale levels. In this regard, the recruitment of RNA-guided genome editing for knocking out miRNAs, flower responsible genes, genes involved in competitive pathways, and base editing are described. The approaches presented here have never been suggested or applied in opium poppy so far.
Keywords: Benzylisoquinoline alkaloids; CRISPR/Cas9; Genome editing; Opium poppy; SNPs; miRNA.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


 Adenine;
Adenine;
 Uracil;
Uracil;
 Cytosine;
Cytosine;
 Guanine
Guanine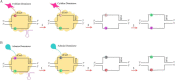
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

