Producing recombinant proteins in Vibrio natriegens
- PMID: 39049057
- PMCID: PMC11267860
- DOI: 10.1186/s12934-024-02455-5
Producing recombinant proteins in Vibrio natriegens
Abstract
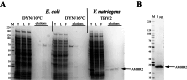
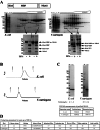
The diversity of chemical and structural attributes of proteins makes it inherently difficult to produce a wide range of proteins in a single recombinant protein production system. The nature of the target proteins themselves, along with cost, ease of use, and speed, are typically cited as major factors to consider in production. Despite a wide variety of alternative expression systems, most recombinant proteins for research and therapeutics are produced in a limited number of systems: Escherichia coli, yeast, insect cells, and the mammalian cell lines HEK293 and CHO. Recent interest in Vibrio natriegens as a new bacterial recombinant protein expression host is due in part to its short doubling time of ≤ 10 min but also stems from the promise of compatibility with techniques and genetic systems developed for E. coli. We successfully incorporated V. natriegens as an additional bacterial expression system for recombinant protein production and report improvements to published protocols as well as new protocols that expand the versatility of the system. While not all proteins benefit from production in V. natriegens, we successfully produced several proteins that were difficult or impossible to produce in E. coli. We also show that in some cases, the increased yield is due to higher levels of properly folded protein. Additionally, we were able to adapt our enhanced isotope incorporation methods for use with V. natriegens. Taken together, these observations and improvements allowed production of proteins for structural biology, biochemistry, assay development, and structure-based drug design in V. natriegens that were impossible and/or unaffordable to produce in E. coli.
Keywords: Escherichia coli; Vibrio natriegens; KRAS4b; TEV protease; auto-induction; isotopic labeling; protein aggregation; protein folding; recombinant protein expression; small GTPase.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

