Comprehensive Analysis of Immune Cell Infiltration and M2-Like Macrophage Biomarker Expression Patterns in Atrial Fibrillation
- PMID: 39049829
- PMCID: PMC11268662
- DOI: 10.2147/IJGM.S462895
Comprehensive Analysis of Immune Cell Infiltration and M2-Like Macrophage Biomarker Expression Patterns in Atrial Fibrillation
Abstract
Background: Macrophages play a crucial role in the progression of AF, closely linked to atrial inflammation and myocardial fibrosis. However, the functions and molecular mechanisms of different phenotypic macrophages in AF are not well understood. This study aims to analyze the infiltration characteristics of atrial immune cells in AF patients and further explore the role and molecular expression patterns of M2 macrophage-related genes in AF.
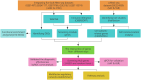
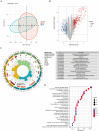
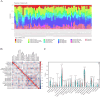
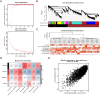
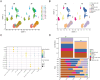
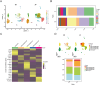
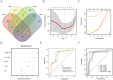
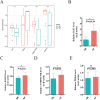
Methods: This study integrates single-cell and large-scale sequencing data to analyze immune cell infiltration and molecular characterization of the LAA in patients with AF, using SR as a control group. CIBERSORT assesses immune cell types in LAA tissues; WGCNA identifies signature genes; cell clustering analyzes cell types and subpopulations; cell communication explores macrophage interactions; hdWGCNA identifies M2 macrophage gene modules in AF. AF biomarkers are identified using LASSO and Random Forest, validated with ROC curves and RT-qPCR. Potential molecular mechanisms are inferred through TF-miRNA-mRNA networks and single-gene enrichment analyses.
Results: Myeloid cell subsets varied considerably between the AF and SR groups, with a significant increase in M2 macrophages in the AF group. Signals of inflammation and matrix remodeling were observed in AF. M2 macrophage-related genes IGF1, PDK4, RAB13, and TMEM176B were identified as AF biomarkers, with RAB13 and TMEM176B being novel markers. A TF-miRNA-mRNA network was constructed using target genes, which are enriched in the PPAR signaling pathway and fatty acid metabolism.
Conclusion: Over infiltration of M2 macrophages may be an important factor in the progression of AF. The M2 macrophage-related genes IGF1, RAB13, TMEM176B and PDK4 may regulate the progression of AF through the PPAR signaling pathway and fatty acid metabolism.
Keywords: M2 macrophage; atrial fibrillation; bulk RNA-sequencing; cardiac fibrosis; inflammation; single-cell RNA-sequencing.
© 2024 Yang et al.
Conflict of interest statement
The authors have no conflicts of interest to declare in this work.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

