Heterochronous multiplex real-time PCR with intercalating dye using uracil-DNA N-glycosylase (UNG) and multiple primer pairs to revaluate post PCR product
- PMID: 39049931
- PMCID: PMC11267051
- DOI: 10.1016/j.mex.2024.102818
Heterochronous multiplex real-time PCR with intercalating dye using uracil-DNA N-glycosylase (UNG) and multiple primer pairs to revaluate post PCR product
Abstract
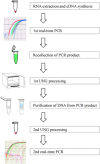
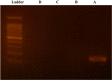
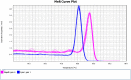
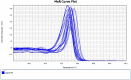
Real-time PCR with intercalating dyes can only be performed once. The expensive fluorescent hydrolysis probes are target specific and are suitable to detect multiplex targets. Uracil-DNA N-glycosylase (UNG), which specifically hydrolyzes and degrades any uracil-containing PCR products, is often applied before PCR to reduce carryover contamination. We developed an optimized protocol for recovering DNA from PCR products and revaluating by real-time PCR with intercalating dye using UNG processing, which is particularly useful when the sample volume is very small and insufficient for multiple assays of real-time PCR.•A real-time PCR master mix with dUTP instead of dTTP was used.•UNG at 1 % and 10 % concentrations of PCR product volumes were used for the first and second processing.•The second real-time PCR was performed with different primer pairs than the first real-time PCR.
Keywords: Heterochronous real-time PCR with intercalating dye using UNG; Multiplex real-time PCR; SYBR green; Uracil-DNA N-glycosylase (UNG).
© 2024 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- Morrison T.B., Weis J.J., Wittwer C.T. Quantification of lowcopy transcripts by continuous SYBR® green I monitoring during amplification. Biotechniques. 1998;24:954–962. http://www.ncbi.nlm.nih.gov/pubmed/9631186 - PubMed
LinkOut - more resources
Full Text Sources

