Is gut microbiota of patients with ALS different from that of healthy individuals?
- PMID: 39050003
- PMCID: PMC11265842
- DOI: 10.55730/1300-0144.5825
Is gut microbiota of patients with ALS different from that of healthy individuals?
Abstract
Background/aim: Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease. Several studies have shown that alterations of microbiota increase the risk of neurodegenerative disorders. We aimed to reveal whether there is a difference in the gut microbiota of patients with ALS.
Materials and methods: The participants are divided into three groups. Group 1 comprised patients with ALS. Healthy family members living in the same house of the patients formed Group 2. Lastly, sex- and age-matched healthy people were included in Group 3. Fecal samples were collected in 15-mL falcon tubes and stored at -80 °C. Genomic DNA isolation was performed on samples. Bacterial primers selected from the 16S rRNA region for the bacterial genome and ITS1 and ITS4 (internal transcribed spacer) were used for the identification of DNA. Next generation sequence analysis (NGS) and taxonomic analyses were performed at the level of bacterial phylum, class, order, family, genus, and species. Alpha and beta diversity indexes were used. The linear discriminant analysis (LDA) effect size method (LEfSe) was applied to identify a microbial taxon specific to ALS disease.
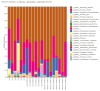
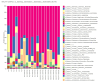
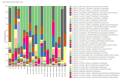
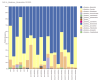
Results: The relative abundances of the Succinivibrionaceae and Lachnospiraceae families were significantly lower in patients. The dominant families among patients were Streptococcaceae and Ruminococcaceae, while the dominant families among healthy controls were Bacteroidaceae and Succinivibrionaceae. The LEfSe analysis revealed that four families (Atopobiaceae, Actinomycetaceae, Erysipelatoclostridiaceae, Peptococcacceae) differed significantly between the patients and healthy controls (LDA values> 2.5, p < 0.05).
Conclusion: Comparison with family members living in the same house is the strength of this study. We found that there were changes in the microbiota of the patients, consistent with the literature. Studies that analyze the composition of the gut microbiota in the predisease period may be needed to understand whether dysbiosis is caused by the mechanisms inherent in the disease or whether it is dysbiosis that initiates the disease.
Keywords: ALS; Amyotrophic lateral sclerosis; gut microbiota; neurodegenerative diseases; pathogenesis of ALS.
© TÜBİTAK.
Conflict of interest statement
Conflicts of interest: The authors declare that they are no conflicts of interest.
Figures



Similar articles
-
Alterations in nasal microbiota of patients with amyotrophic lateral sclerosis.Chin Med J (Engl). 2024 Jan 20;137(2):162-171. doi: 10.1097/CM9.0000000000002701. Epub 2023 Jul 21. Chin Med J (Engl). 2024. PMID: 37482646 Free PMC article.
-
The fecal microbiome of ALS patients.Neurobiol Aging. 2018 Jan;61:132-137. doi: 10.1016/j.neurobiolaging.2017.09.023. Epub 2017 Oct 3. Neurobiol Aging. 2018. PMID: 29065369
-
The alteration of gut microbiome and metabolism in amyotrophic lateral sclerosis patients.Sci Rep. 2020 Aug 3;10(1):12998. doi: 10.1038/s41598-020-69845-8. Sci Rep. 2020. PMID: 32747678 Free PMC article.
-
Potential Role of the Gut Microbiome in ALS: A Systematic Review.Biol Res Nurs. 2018 Oct;20(5):513-521. doi: 10.1177/1099800418784202. Epub 2018 Jun 20. Biol Res Nurs. 2018. PMID: 29925252
-
Gut microbiota in ALS: possible role in pathogenesis?Expert Rev Neurother. 2019 Sep;19(9):785-805. doi: 10.1080/14737175.2019.1623026. Epub 2019 May 29. Expert Rev Neurother. 2019. PMID: 31122082 Review.
Cited by
-
The association between gut microbiota composition and cardiometabolic parameters in healthy adults.BMC Microbiol. 2025 Aug 14;25(1):505. doi: 10.1186/s12866-025-04261-4. BMC Microbiol. 2025. PMID: 40813961 Free PMC article.
-
Gut microbiota immune cross-talk in amyotrophic lateral sclerosis.Neurotherapeutics. 2024 Oct;21(6):e00469. doi: 10.1016/j.neurot.2024.e00469. Epub 2024 Nov 6. Neurotherapeutics. 2024. PMID: 39510899 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

