Relationships between systemic sclerosis and atherosclerosis: screening for mitochondria-related biomarkers
- PMID: 39050259
- PMCID: PMC11266065
- DOI: 10.3389/fgene.2024.1375331
Relationships between systemic sclerosis and atherosclerosis: screening for mitochondria-related biomarkers
Abstract
Background: Patients with systemic sclerosis (SSc) are known to have higher incidence of atherosclerosis (AS). Mitochondrial injuries in SSc can cause endothelial dysfunction, leading to AS; thus, mitochondria appear to be hubs linking SSc to AS. This study aimed to identify the mitochondria-related biomarkers of SSc and AS.
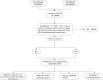
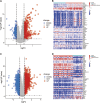
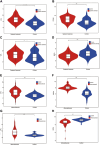
Methods: We identified common differentially expressed genes (DEGs) in the SSc (GSE58095) and AS (GSE100927) datasets of the Gene Expression Omnibus (GEO) database. Considering the intersection between genes with identical expression trends and mitochondrial genes, we used the least absolute shrinkage and selection operator (LASSO) as well as random forest (RF) algorithms to identify four mitochondria-related hub genes. Diagnostic nomograms were then constructed to predict the likelihood of SSc and AS. Next, we used the CIBERSORT algorithm to evaluate immune infiltration in both disorders, predicted the transcription factors for the hub genes, and validated these genes for the two datasets.
Results: A total of 112 genes and 13 mitochondria-related genes were identified; these genes were then significantly enriched for macrophage differentiation, collagen-containing extracellular matrix, collagen binding, antigen processing and presentation, leukocyte transendothelial migration, and apoptosis. Four mitochondria-related hub DEGs (IFI6, FSCN1, GAL, and SGCA) were also identified. The nomograms showed good diagnostic values for GSE58095 (area under the curve (AUC) = 0.903) and GSE100927 (AUC = 0.904). Further, memory B cells, γδT cells, M0 macrophages, and activated mast cells were significantly higher in AS, while the resting memory CD4+ T cells were lower and M1 macrophages were higher in SSc; all of these were closely linked to multiple immune cells. Gene set enrichment analysis (GSEA) showed that IFI6 and FSCN1 were involved in immune-related pathways in both AS and SSc; GAL and SGCA are related to mitochondrial metabolism pathways in both SSc and AS. Twenty transcription factors (TFs) were predicted, where two TFs, namely BRCA1 and PPARγ, were highly expressed in both SSc and AS.
Conclusion: Four mitochondria-related biomarkers were identified in both SSc and AS, which have high diagnostic value and are associated with immune cell infiltration in both disorders. Hence, this study provides new insights into the pathological mechanisms underlying SSc and AS. The specific roles and action mechanisms of these genes require further clinical validation in SSc patients with AS.
Keywords: atherosclerosis; immune infiltration; machine learning; mitochondria-related genes; systemic sclerosis.
Copyright © 2024 Wang, Lyu, Qin and Xie.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

