Population genomics and distribution modeling revealed the history and suggested a possible future of the endemic Agave aurea (Asparagaceae) complex in the Baja California Peninsula
- PMID: 39050658
- PMCID: PMC11267983
- DOI: 10.1002/ece3.70027
Population genomics and distribution modeling revealed the history and suggested a possible future of the endemic Agave aurea (Asparagaceae) complex in the Baja California Peninsula
Abstract
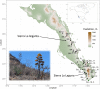
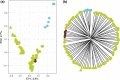
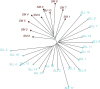
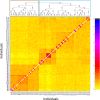
Agaves are an outstanding arid-adapted group of species that provide a unique chance to study the influence of multiple potential factors (i.e., geological and ecological) on plant population structure and diversification in the heterogeneous environment of the Baja California Peninsula. However, relatively little is known about the phylogeography of the endemic agave species of this region. Herein, we used over 10,000 single-nucleotide polymorphisms (SNPs) and spatial data from the Agave aurea species complex (i.e., A. aurea ssp. aurea, A. aurea ssp. promontorii, and A. aurea var. capensis) to resolve genetic relationships within this complex and uncover fine-scale population structure, diversity patterns, and their potential underlying drivers. Analyses resolved low genetic structure within this complex, suggesting that A. aurea is more likely to represent several closely related populations than separate species or varieties/subspecies. We found that geographical and historical ecological characteristics-including precipitation, latitude, and past climatic fluctuations-have played an important role in the spatial distribution of diversity and structure in A. aurea. Finally, species distribution modeling results suggested that climate change will become critical in the extinction risk of A. aurea, with the northernmost population being particularly vulnerable. The low population genetic structure found in A. aurea is consistent with agave's life history, and it is probably related to continuity of distribution, relatively low habitat fragmentation, and dispersion by pollinators. Together, these findings have important implications for management and conservation programs in agave, such as creating and evaluating protected areas and translocating and augmentation of particular populations.
Keywords: Agavoideae; Baja California Peninsula; Sonoran Desert; climate change; genomic diversity; pollinators.
© 2024 The Author(s). Ecology and Evolution published by John Wiley & Sons Ltd.
Conflict of interest statement
None declared.
Figures



References
-
- Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data [Online]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Associated data
LinkOut - more resources
Full Text Sources
Miscellaneous

