Limb connective tissue is organized in a continuum of promiscuous fibroblast identities during development
- PMID: 39050702
- PMCID: PMC11267076
- DOI: 10.1016/j.isci.2024.110305
Limb connective tissue is organized in a continuum of promiscuous fibroblast identities during development
Abstract
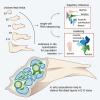
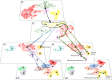
Connective tissue (CT), which includes tendon and muscle CT, plays critical roles in development, in particular as positional cue provider. Nonetheless, our understanding of fibroblast developmental programs is hampered because fibroblasts are highly heterogeneous and poorly characterized. Combining single-cell RNA-sequencing-based strategies including trajectory inference and in situ hybridization analyses, we address the diversity of fibroblasts and their developmental trajectories during chicken limb fetal development. We show that fibroblasts switch from a positional information to a lineage diversification program at the fetal period onset. Muscle CT and tendon are composed of several fibroblast populations that emerge asynchronously. Once the final muscle pattern is set, transcriptionally close populations are found in neighboring locations in limbs, prefiguring the adult fibroblast layers. We propose that the limb CT is organized in a continuum of promiscuous fibroblast identities, allowing for the robust and efficient connection of muscle to bone and skin.
Keywords: Biological sciences; Cell biology; Developmental biology; Molecular biology.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

