Development of multiplex real-time PCR for simultaneous detection of SARS-CoV-2, CCoV, and FIPV
- PMID: 39051010
- PMCID: PMC11266814
- DOI: 10.3389/fvets.2024.1337690
Development of multiplex real-time PCR for simultaneous detection of SARS-CoV-2, CCoV, and FIPV
Abstract
Introduction: Coronaviruses, including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), canine coronavirus (CCoV), and feline infectious peritonitis virus (FIPV), have the potential for interspecies transmission. These viruses can be present in complex environments where humans, dogs, and cats coexist, posing a significant threat to both human and animal safety.
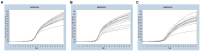
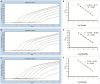
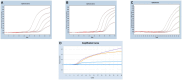
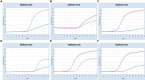
Methods and results: In this study, we developed a novel multiplex TaqMan-probe-based real-time PCR assay for the simultaneous detection and differentiation of SARS-CoV-2, CCoV, and FIPV. Specific primers and TaqMan fluorescent probes were designed based on the N region of SARS-CoV-2 and FIPV, as well as the S region of CCoV, which demonstrated a remarkable sensitivity and specificity toward the targeted viruses, as few as 21.83, 17.25 and 9.25 copies/μL for SARS-CoV-2, CCoV and FIPV, respectively. The standard curve constructed by the optimized method in our present study showed a high amplification efficiency within or near the optimal range of 91% to 116% and R(2) values were at least 0.95 for the abovementioned coronaviruses. A total of 91 samples, including six plasmid mixed mock samples, four virus fluid mixing simulated samples, and 81 clinical samples, were analyzed using this method. Results demonstrated strong agreement with conventional approaches.
Discussion: By enabling the simultaneous detection of three viruses, this method enhances testing efficiency while decreasing costs. Importantly, it provides a valuable tool for the prevalence and geographical distribution of suspected and co-infected animals, ultimately contributing to the advancement of both animal and public health.
Keywords: FIPV; SARS-CoV-2; canine coronavirus; detection; real-time PCR.
Copyright © 2024 Liu, Zhu, Du, Zhu, Pan, Yin and Sun.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Phylogenetic analysis of feline infectious peritonitis virus, feline enteric coronavirus, and severe acute respiratory syndrome coronavirus 2 of cats in Surabaya, Indonesia.Vet World. 2023 Jan;16(1):76-81. doi: 10.14202/vetworld.2023.76-81. Epub 2023 Jan 11. Vet World. 2023. PMID: 36855370 Free PMC article.
-
Infection of cats with atypical feline coronaviruses harbouring a truncated form of the canine type I non-structural ORF3 gene.Infect Genet Evol. 2013 Dec;20:488-94. doi: 10.1016/j.meegid.2013.09.024. Epub 2013 Oct 9. Infect Genet Evol. 2013. PMID: 24121017 Free PMC article.
-
Development of a TaqMan-probe-based multiplex real-time PCR for the simultaneous detection of emerging and reemerging swine coronaviruses.Virulence. 2020 Jan 1;11(1):707-718. doi: 10.1080/21505594.2020.1771980. Virulence. 2020. PMID: 32490723 Free PMC article.
-
Current concepts in the development of therapeutics against human and animal coronavirus diseases by targeting NP.Comput Struct Biotechnol J. 2021;19:1072-1080. doi: 10.1016/j.csbj.2021.01.032. Epub 2021 Jan 30. Comput Struct Biotechnol J. 2021. PMID: 33552444 Free PMC article. Review.
-
The knotty biology of canine coronavirus: A worrying model of coronaviruses' danger.Res Vet Sci. 2022 May;144:190-195. doi: 10.1016/j.rvsc.2021.11.014. Epub 2021 Nov 20. Res Vet Sci. 2022. PMID: 34838321 Free PMC article. Review.
Cited by
-
The quadruplex TaqMan MGB fluorescent quantitative PCR method for simultaneous detection of feline panleukopenia virus, feline herpesvirus 1, feline calicivirus and feline infectious peritonitis virus.Front Cell Infect Microbiol. 2025 May 30;15:1581946. doi: 10.3389/fcimb.2025.1581946. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40521026 Free PMC article.
References
-
- Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, et al. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. (2012) 86:3995–4008. doi: 10.1128/JVI.06540-11, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

