NK Cytotoxicity Mediated by NK-92 Cell Lines Expressing Combinations of Two Allelic Variants for FCGR3
- PMID: 39051331
- PMCID: PMC11270249
- DOI: 10.3390/antib13030055
NK Cytotoxicity Mediated by NK-92 Cell Lines Expressing Combinations of Two Allelic Variants for FCGR3
Abstract
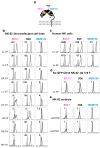
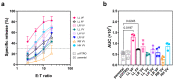
Natural killer (NK) cells play an important role in the surveillance of viral infections and cancer. NK cell antibody-dependent cellular cytotoxicity (ADCC) and direct cytotoxicity are mediated by the recognition of antibody-coated target cells through the Fc gamma receptor IIIA (FcγRIIIa/CD16) and by ligands of activating/inhibitory NK receptors, respectively. Allelic variants of the FCGR3A gene include the high-affinity single-nucleotide polymorphism (SNP) rs396991 (V176F), which is associated with the efficacy of monoclonal antibody (mAb) therapies, and the SNP rs10127939 (L66H/R). The contribution of FCGR3A SNPs to NK cell effector functions remains controversial; therefore, we generated a panel of eight NK-92 cell lines expressing specific combinations of these SNPs and tested their cytotoxicities. NK-92 cells were stably transfected with plasmids containing different combinations of FCGR3A SNPs. Messenger RNA and FcγRIIIa/CD16 cell surface expressions were detected using new generation sequencing (NGS) and flow cytometry, respectively. All FcγRIIIa/CD16-transfected NK-92 cell lines exhibited robust ADCC against three different target cell lines with minor differences. In addition, enhanced direct NK cytotoxicity against K562 target cells was observed, suggesting a mechanistic role of FcγRIIIa/CD16 in direct NK cytotoxicity. In conclusion, we generated eight FcγRIIIa/CD16-transfected NK-92 cell lines carrying different combinations of two of the most studied FCGR3A SNPs, representing the major genotypes described in the European population. The functional characterization of these cell lines revealed differences in ADCC and direct NK cytotoxicity that may have implications for the design of adoptive cancer immunotherapies using NK cells and tumor antigen-directed mAbs.
Keywords: ADCC; CD16; FCGR3A; L48H/R; L66H/R; NK-92; V158F; V176F.
Conflict of interest statement
The authors declare no conflicts of interest except for M.P. and G.L.P.Y., whose salaries were partially financed by CSL Behring. The funder had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures





Similar articles
-
The influence of NK cell-mediated ADCC: Structure and expression of the CD16 molecule differ among FcγRIIIa-V158F genotypes in healthy Japanese subjects.Hum Immunol. 2016 Feb;77(2):165-71. doi: 10.1016/j.humimm.2015.11.001. Epub 2015 Nov 12. Hum Immunol. 2016. PMID: 26582002
-
Anti-tumor effects of NK cells and anti-PD-L1 antibody with antibody-dependent cellular cytotoxicity in PD-L1-positive cancer cell lines.J Immunother Cancer. 2020 Aug;8(2):e000873. doi: 10.1136/jitc-2020-000873. J Immunother Cancer. 2020. PMID: 32830112 Free PMC article.
-
SNPs rs4656317 and rs12071048 located within an enhancer in FCGR3A are in strong linkage disequilibrium with rs396991 and influence NK cell-mediated ADCC by transcriptional regulation.Hum Immunol. 2016 Oct;77(10):997-1003. doi: 10.1016/j.humimm.2016.06.012. Epub 2016 Jun 20. Hum Immunol. 2016. PMID: 27338556
-
Natural Killer (NK) Cell Education Differentially Influences HIV Antibody-Dependent NK Cell Activation and Antibody-Dependent Cellular Cytotoxicity.Front Immunol. 2017 Aug 24;8:1033. doi: 10.3389/fimmu.2017.01033. eCollection 2017. Front Immunol. 2017. PMID: 28883824 Free PMC article. Review.
-
Potentiation of natural killer cells to overcome cancer resistance to NK cell-based therapy and to enhance antibody-based immunotherapy.Front Immunol. 2023 Nov 24;14:1275904. doi: 10.3389/fimmu.2023.1275904. eCollection 2023. Front Immunol. 2023. PMID: 38077389 Free PMC article. Review.
Cited by
-
Fc gamma receptor polymorphisms in antibody therapy: implications for bioassay development to enhance product quality.Antib Ther. 2025 Jan 21;8(2):87-98. doi: 10.1093/abt/tbaf003. eCollection 2025 Apr. Antib Ther. 2025. PMID: 40177643 Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

