Integrated Analysis of Transcriptome Profiles and lncRNA-miRNA-mRNA Competing Endogenous RNA Regulatory Network to Identify Biological Functional Effects of Genes and Pathways Associated with Johne's Disease in Dairy Cattle
- PMID: 39051372
- PMCID: PMC11270299
- DOI: 10.3390/ncrna10040038
Integrated Analysis of Transcriptome Profiles and lncRNA-miRNA-mRNA Competing Endogenous RNA Regulatory Network to Identify Biological Functional Effects of Genes and Pathways Associated with Johne's Disease in Dairy Cattle
Abstract
Paratuberculosis or Johne's disease (JD), a chronic granulomatous gastroenteritis caused by Mycobacterium avium subsp. paratuberculosis (MAP), causes huge economic losses and reduces animal welfare in dairy cattle herds worldwide. At present, molecular mechanisms and biological functions involved in immune responses to MAP infection of dairy cattle are not clearly understood. Our purpose was to integrate transcriptomic profiles and competing endogenous RNA (ceRNA) network analyses to identify key messenger RNAs (mRNAs) and regulatory RNAs involved in molecular regulation of peripheral blood mononuclear cells (PBMCs) for MAP infection in dairy cattle. In total, 28 lncRNAs, 42 miRNAs, and 370 mRNAs were identified by integrating gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses. In this regard, we identified 21 hub genes (CCL20, CCL5, CD40, CSF2, CXCL8, EIF2AK2, FOS, IL10, IL17A, IL1A, IL1B, IRF1, MX2, NFKB1, NFKBIA, PTGS2, SOCS3, TLR4, TNF, TNFAIP3, and VCAM1) involved in MAP infection. Furthermore, eight candidate subnets with eight lncRNAs, 29 miRNAs, and 237 mRNAs were detected through clustering analyses, whereas GO enrichment analysis of identified RNAs revealed 510, 22, and 11 significantly enriched GO terms related to MAP infection in biological process, molecular function, and cellular component categories, respectively. The main metabolic-signaling pathways related to MAP infection that were enriched included the immune system process, defense response, response to cytokine, leukocyte migration, regulation of T cell activation, defense response to bacterium, NOD-like receptor, B cell receptor, TNF, NF-kappa B, IL-17, and T cell receptor signaling pathways. Contributions of transcriptome profiles from MAP-positive and MAP-negative sample groups plus a ceRNA regulatory network underlying phenotypic differences in the intensity of pathogenicity of JD provided novel insights into molecular mechanisms associated with immune system responses to MAP infection in dairy cattle.
Keywords: MAP infection; ceRNA regulatory network; dairy cattle; immune responses; paratuberculosis; transcriptome analysis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
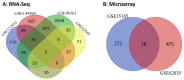
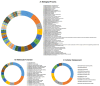
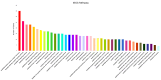


References
-
- Marete A., Ariel O., Ibeagha-Awemu E., Bissonnette N. Identification of long non-coding RNA isolated from naturally infected macrophages and associated with bovine Johne’s Disease in Canadian Holstein using a combination of neural networks and logistic regression. Front. Vet. Sci. 2021;8:639053. doi: 10.3389/fvets.2021.639053. - DOI - PMC - PubMed
-
- Malvisi M., Curti N., Remondini D., De Iorio M.G., Palazzo F., Gandini G., Vitali S., Polli M., Williams J.L., Minozzi G. Combinatorial discriminant analysis applied to RNAseq data reveals a set of 10 transcripts as signatures of exposure of cattle to Mycobacterium avium subsp. paratuberculosis. Animals. 2020;10:253. doi: 10.3390/ani10020253. - DOI - PMC - PubMed
-
- Park H.T., Park H.E., Shim S., Kim S., Shin M.K., Yoo H.S. Epithelial processed Mycobacterium avium subsp. paratuberculosis induced prolonged Th17 response and suppression of phagocytic maturation in bovine peripheral blood mononuclear cells. Sci. Rep. 2020;10:21048. doi: 10.1038/s41598-020-78113-8. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

